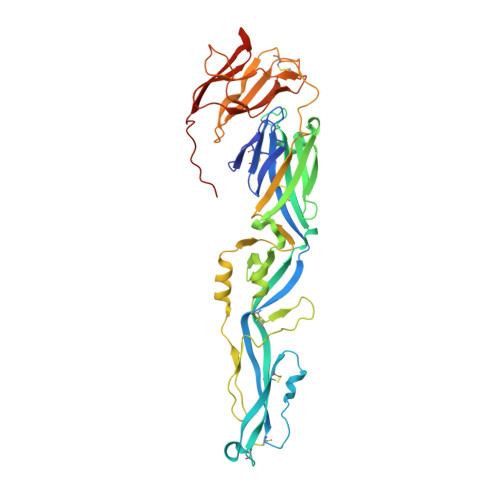
Crystal structure of Usutu virus envelope protein in the pre-fusion state
Chen, Z., Ye, F., Lin, S., Yang, F., Cheng, Y., Cao, Y., Chen, Z., Lu, G.(2018) Virol J 15: 183-183
- PubMed: 30477514
- DOI: https://doi.org/10.1186/s12985-018-1092-6
- Primary Citation of Related Structures:
6A0P - PubMed Abstract:
Usutu virus (USUV) is a mosquito-born flavivirus that can infect multiple avian and mammalian species. The viral surface envelope (E) protein functions to initiate the viral infection by recognizing cellular receptors and mediating the subsequent membrane fusion, and is therefore a key virulence factor involved in the pathogenesis of USUV. The structural features of USUV-E, however, remains un-investigated thus far. Using the crystallographic method, we determined the structure of USUV-E in the pre-fusion state at 2.0 angstrom. As expected, the overall fold of USUV-E, with three β-barrel domains (DI, DII, and DIII), resembles those of other flaviviral E proteins. In comparison to other pre-fusion E structures, however, USUV-E exhibits an apparently enlarged inter-domain angle between DI and DII, leading to a more extended conformation. Using our structure and other reported pre-fusion E structures, the DI-DII domain-angle difference was analyzed in a pairwise manner. The result shows a much higher degree of variations for USUV-E, indicating the potential for remarkable DI-DII domain angle plasticity among flaviviruses. We report the crystal structure of USUV-E and show that its pre-fusion structure has an enlarged DI-DII domain-angle which has not been observed in other reported flaviviral E-structures.
Organizational Affiliation:
West China Hospital Emergency Department (WCHED), State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, Chengdu, 610041, Sichuan, China.