Alkynyl Benzoxazines and Dihydroquinazolines as Cysteine Targeting Covalent Warheads and Their Application in Identification of Selective Irreversible Kinase Inhibitors.
McAulay, K., Hoyt, E.A., Thomas, M., Schimpl, M., Bodnarchuk, M.S., Lewis, H.J., Barratt, D., Bhavsar, D., Robinson, D.M., Deery, M.J., Ogg, D.J., Bernardes, G.J.L., Ward, R.A., Waring, M.J., Kettle, J.G.(2020) J Am Chem Soc 142: 10358-10372
- PubMed: 32412754
- DOI: https://doi.org/10.1021/jacs.9b13391
- Primary Citation of Related Structures:
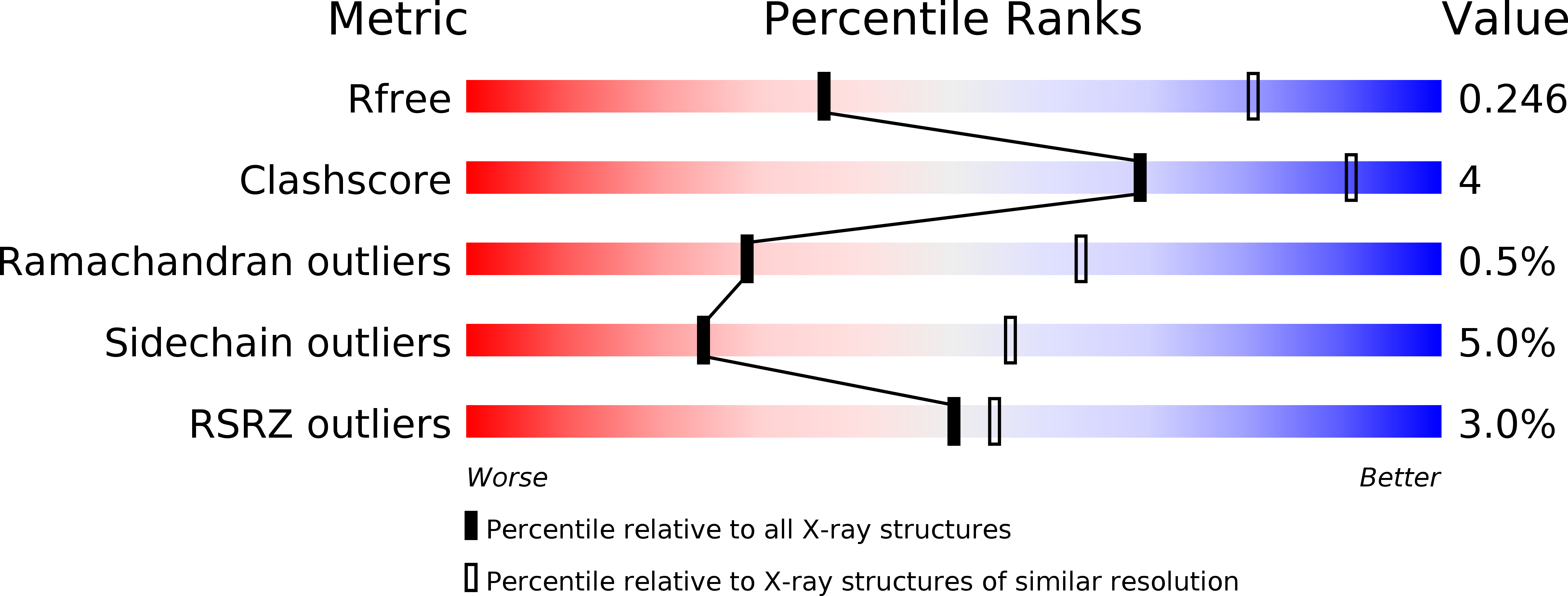
6XV9, 6XVA, 6XVB, 6XVJ, 6XVK - PubMed Abstract:
With a resurgence in interest in covalent drugs, there is a need to identify new moieties capable of cysteine bond formation that are differentiated from commonly employed systems such as acrylamide. Herein, we report on the discovery of new alkynyl benzoxazine and dihydroquinazoline moieties capable of covalent reaction with cysteine. Their utility as alternative electrophilic warheads for chemical biological probes and drug molecules is demonstrated through site-selective protein modification and incorporation into kinase drug scaffolds. A potent covalent inhibitor of JAK3 kinase was identified with superior selectivity across the kinome and improvements in in vitro pharmacokinetic profile relative to the related acrylamide-based inhibitor. In addition, the use of a novel heterocycle as a cysteine reactive warhead is employed to target Cys788 in c-KIT, where acrylamide has previously failed to form covalent interactions. These new reactive and selective heterocyclic warheads supplement the current repertoire for cysteine covalent modification while avoiding some of the limitations generally associated with established moieties.
Organizational Affiliation:
Oncology R&D, AstraZeneca, Cambridge CB4 0WG, U.K.