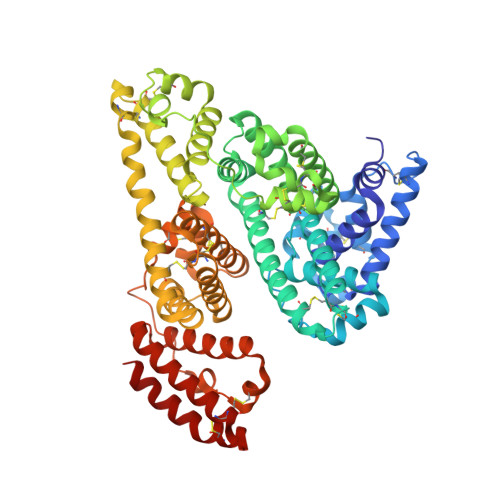
Molecular determinants of vascular transport of dexamethasone in COVID-19 therapy.
Shabalin, I.G., Czub, M.P., Majorek, K.A., Brzezinski, D., Grabowski, M., Cooper, D.R., Panasiuk, M., Chruszcz, M., Minor, W.(2020) IUCrJ 7
- PubMed: 33063792
- DOI: https://doi.org/10.1107/S2052252520012944
- Primary Citation of Related Structures:
6XK0 - PubMed Abstract:
Dexamethasone, a widely used corticosteroid, has recently been reported as the first drug to increase the survival chances of patients with severe COVID-19. Therapeutic agents, including dexamethasone, are mostly transported through the body by binding to serum albumin. Here, the first structure of serum albumin in complex with dexamethasone is reported. Dexamethasone binds to drug site 7, which is also the binding site for commonly used nonsteroidal anti-inflammatory drugs and testosterone, suggesting potentially problematic binding competition. This study bridges structural findings with an analysis of publicly available clinical data from Wuhan and suggests that an adjustment of the dexamethasone regimen should be further investigated as a strategy for patients affected by two major COVID-19 risk factors: low albumin levels and diabetes.
Organizational Affiliation:
Department of Molecular Physiology and Biological Physics, University of Virginia, 1340 Jefferson Park Avenue, Charlottesville, VA 22908, USA.