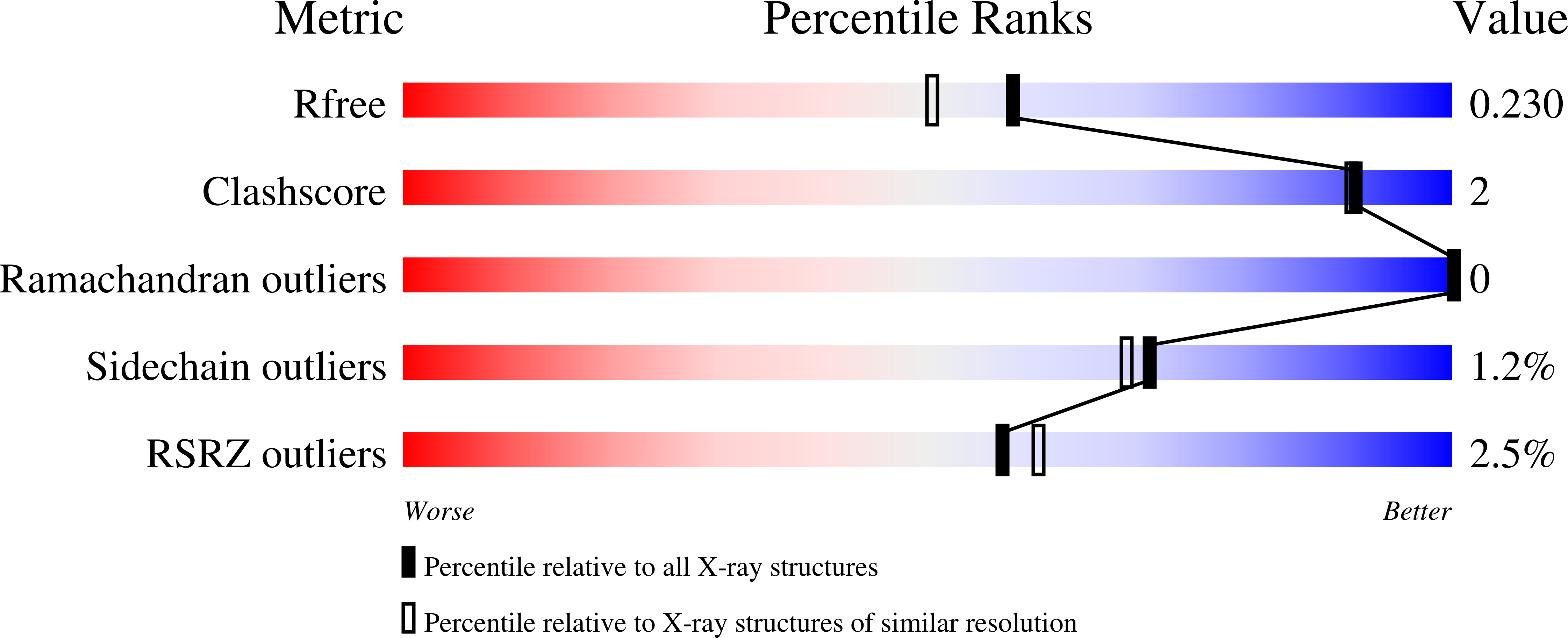
Post-translational Succinylation ofMycobacterium tuberculosisEnoyl-CoA Hydratase EchA19 Slows Catalytic Hydration of Cholesterol Catabolite 3-Oxo-chol-4,22-diene-24-oyl-CoA.
Bonds, A.C., Yuan, T., Werman, J.M., Jang, J., Lu, R., Nesbitt, N.M., Garcia-Diaz, M., Sampson, N.S.(2020) ACS Infect Dis 6: 2214-2224
- PubMed: 32649175
- DOI: https://doi.org/10.1021/acsinfecdis.0c00329
- Primary Citation of Related Structures:
6WYI - PubMed Abstract:
Cholesterol is a major carbon source for Mycobacterium tuberculosis ( Mtb ) during infection, and cholesterol utilization plays a significant role in persistence and virulence within host macrophages. Elucidating the mechanism by which cholesterol is degraded may permit the identification of new therapeutic targets. Here, we characterized EchA19 (Rv3516), an enoyl-CoA hydratase involved in cholesterol side-chain catabolism. Steady-state kinetics assays demonstrated that EchA19 preferentially hydrates cholesterol enoyl-CoA metabolite 3-oxo-chol-4,22-diene-24-oyl-CoA, an intermediate of side-chain β-oxidation. In addition, succinyl-CoA, a downstream catabolite of propionyl-CoA that forms during cholesterol degradation, covalently modifies targeted mycobacterial proteins, including EchA19. Inspection of a 1.9 Å resolution X-ray crystallography structure of Mtb EchA19 suggests that succinylation of Lys132 and Lys139 may perturb enzymatic activity by modifying the entrance to the substrate binding site. Treatment of EchA19 with succinyl-CoA revealed that these two residues are hotspots for succinylation. Replacement of these specific lysine residues with negatively charged glutamate reduced the rate of catalytic hydration of 3-oxo-chol-4,22-diene-24-oyl-CoA by EchA19, as does succinylation of EchA19. Our findings suggest that succinylation is a negative feedback regulator of cholesterol metabolism, thereby adding another layer of complexity to Mtb physiology in the host. These regulatory pathways are potential noncatabolic targets for antimicrobial drugs.
Organizational Affiliation:
Department of Pharmacological Sciences, Stony Brook University, Stony Brook, New York 11794-8651.