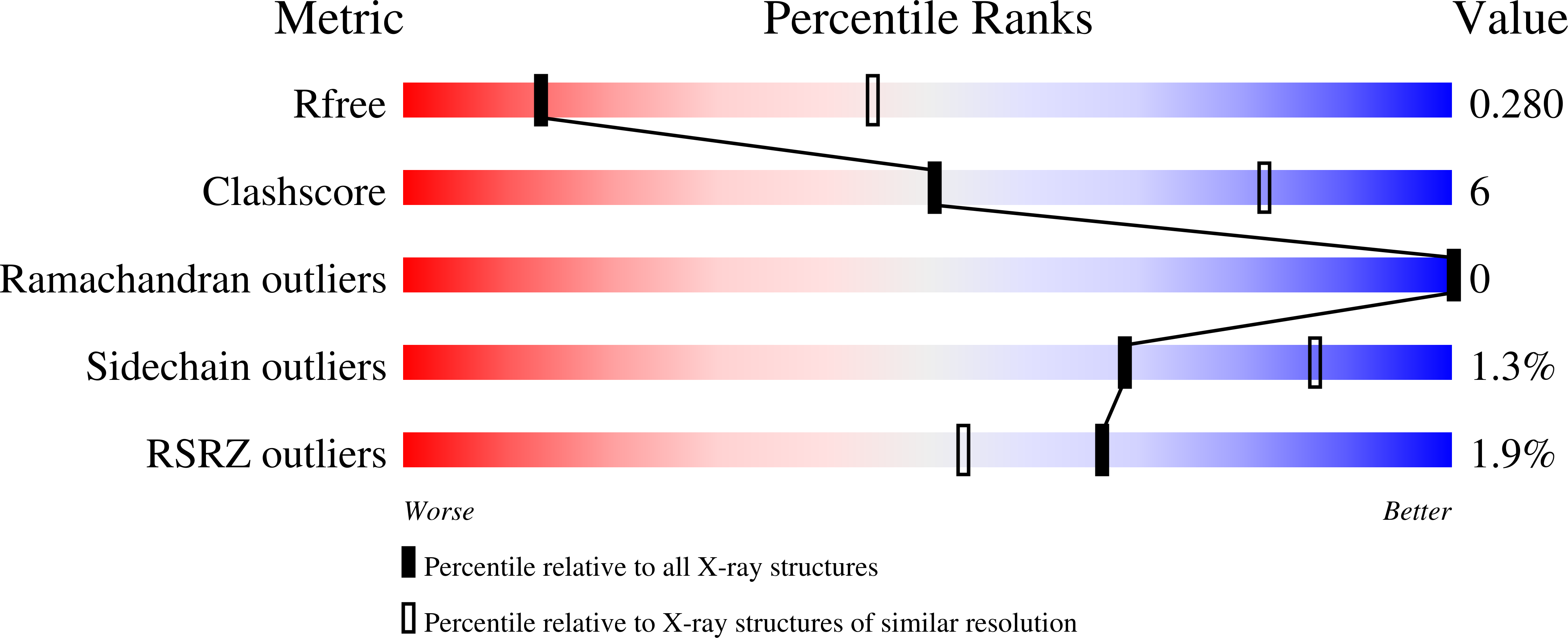
The RING Domain of RING Finger 12 Efficiently Builds Degradative Ubiquitin Chains.
Middleton, A.J., Zhu, J., Day, C.L.(2020) J Mol Biol 432: 3790-3801
- PubMed: 32416094
- DOI: https://doi.org/10.1016/j.jmb.2020.05.001
- Primary Citation of Related Structures:
6W7Z, 6W9A, 6W9D - PubMed Abstract:
RNF12 is a widely expressed ubiquitin E3 ligase that is required for X-chromosome inactivation, regulation of LIM-domain containing transcription factors, and TGF-β signaling. A RING domain at the C terminus of RNF12 is important for its E3 ligase activity, and mutations in the RING domain are associated with X-linked intellectual disability. Here we have characterized ubiquitin transfer by RNF12, and show that the RING domain can bind to, and is active with, ubiquitin conjugating enzymes (E2s) that produce degradative ubiquitin chains. We report the crystal structures of RNF12 in complex with two of these E2 enzymes, as well as with an E2~Ub conjugate in a closed conformation. These structures form a basis for understanding the deleterious effect of a number of disease causing mutations. Comparison of the RNF12 structure with other monomeric RINGs suggests that a loop prior to the core RING domain has a conserved and essential role in stabilization of the active conformation of the bound E2~Ub conjugate. Together these findings provide a framework for better understanding substrate ubiquitylation by RNF12 and the impact of disease causing mutations.
Organizational Affiliation:
Department of Biochemistry, School of Biomedical Sciences, University of Otago, Dunedin 9054, New Zealand.