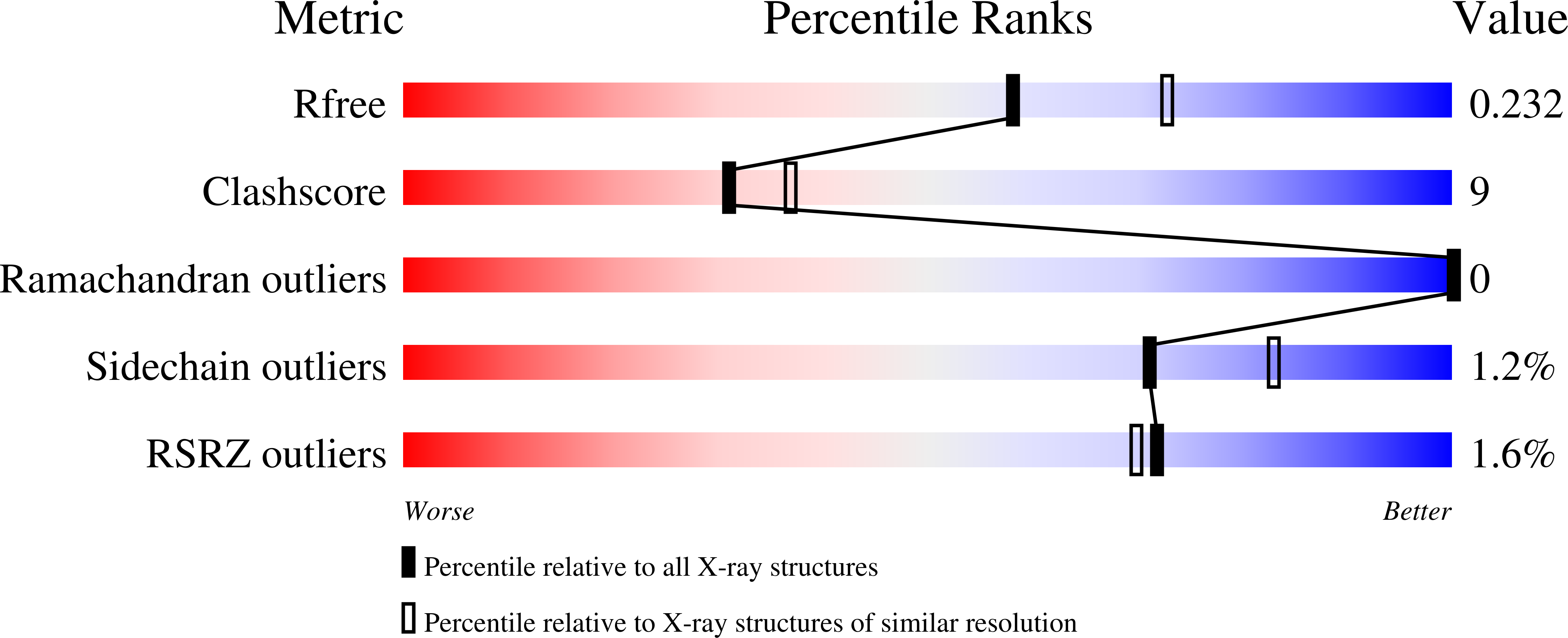
Structure of the Peptidoglycan Synthase Activator LpoP in Pseudomonas aeruginosa.
Caveney, N.A., Egan, A.J.F., Ayala, I., Laguri, C., Robb, C.S., Breukink, E., Vollmer, W., Strynadka, N.C.J., Simorre, J.P.(2020) Structure 28: 643-650.e5
- PubMed: 32320673
- DOI: https://doi.org/10.1016/j.str.2020.03.012
- Primary Citation of Related Structures:
6W5Q - PubMed Abstract:
Peptidoglycan (PG) is an essential component of the bacterial cell wall and is assembled from a lipid II precursor by glycosyltransferase and transpeptidase reactions catalyzed in particular by bifunctional class A penicillin-binding proteins (aPBPs). In the major clinical pathogen Pseudomonas aeruginosa, PBP1B is anchored within the cytoplasmic membrane but regulated by a bespoke outer membrane-localized lipoprotein known as LpoP. Here, we report the structure of LpoP, showing an extended N-terminal, flexible tether followed by a well-ordered C-terminal tandem-tetratricopeptide repeat domain. We show that LpoP stimulates both PBP1B transpeptidase and glycosyltransferase activities in vitro and interacts directly via its C terminus globular domain with the central UB2H domain of PBP1B. Contrary to the situation in E. coli, P. aeruginosa CpoB does not regulate PBP1B/LpoP in vitro. We propose a mechanism that helps to underscore similarities and differences in class A PBP activation across Gram-negative bacteria.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology and Centre for Blood Research, The University of British Columbia, Vancouver V6T 1Z3 BC, Canada.