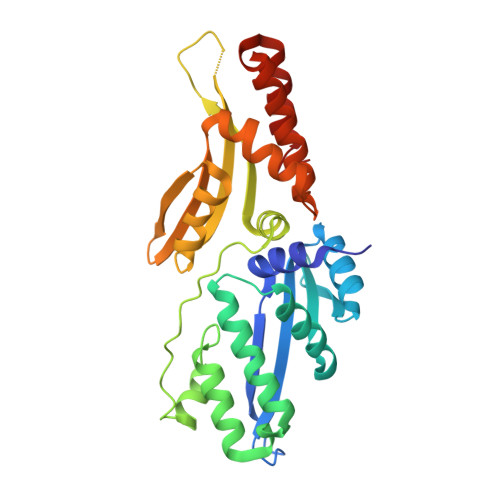
Optimization of Protein Thermostability and Exploitation of Recognition Behavior to Engineer Altered Protein-DNA Recognition.
Lambert, A.R., Hallinan, J.P., Werther, R., Glow, D., Stoddard, B.L.(2020) Structure 28: 760-775.e8
- PubMed: 32359399
- DOI: https://doi.org/10.1016/j.str.2020.04.009
- Primary Citation of Related Structures:
6UVW, 6UW0, 6UWG, 6UWH, 6UWJ, 6UWK - PubMed Abstract:
The redesign of a macromolecular binding interface and corresponding alteration of recognition specificity is a challenging endeavor that remains recalcitrant to computational approaches. This is particularly true for the redesign of DNA binding specificity, which is highly dependent upon bending, hydrogen bonds, electrostatic contacts, and the presence of solvent and counterions throughout the molecular interface. Thus, redesign of protein-DNA binding specificity generally requires iterative rounds of amino acid randomization coupled to selections. Here, we describe the importance of scaffold thermostability for protein engineering, coupled with a strategy that exploits the protein's specificity profile, to redesign the specificity of a pair of meganucleases toward three separate genomic targets. We determine and describe a series of changes in protein sequence, stability, structure, and activity that accumulate during the engineering process, culminating in fully retargeted endonucleases.
Organizational Affiliation:
Division of Basic Sciences, Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue N., Seattle, WA 98109, USA.