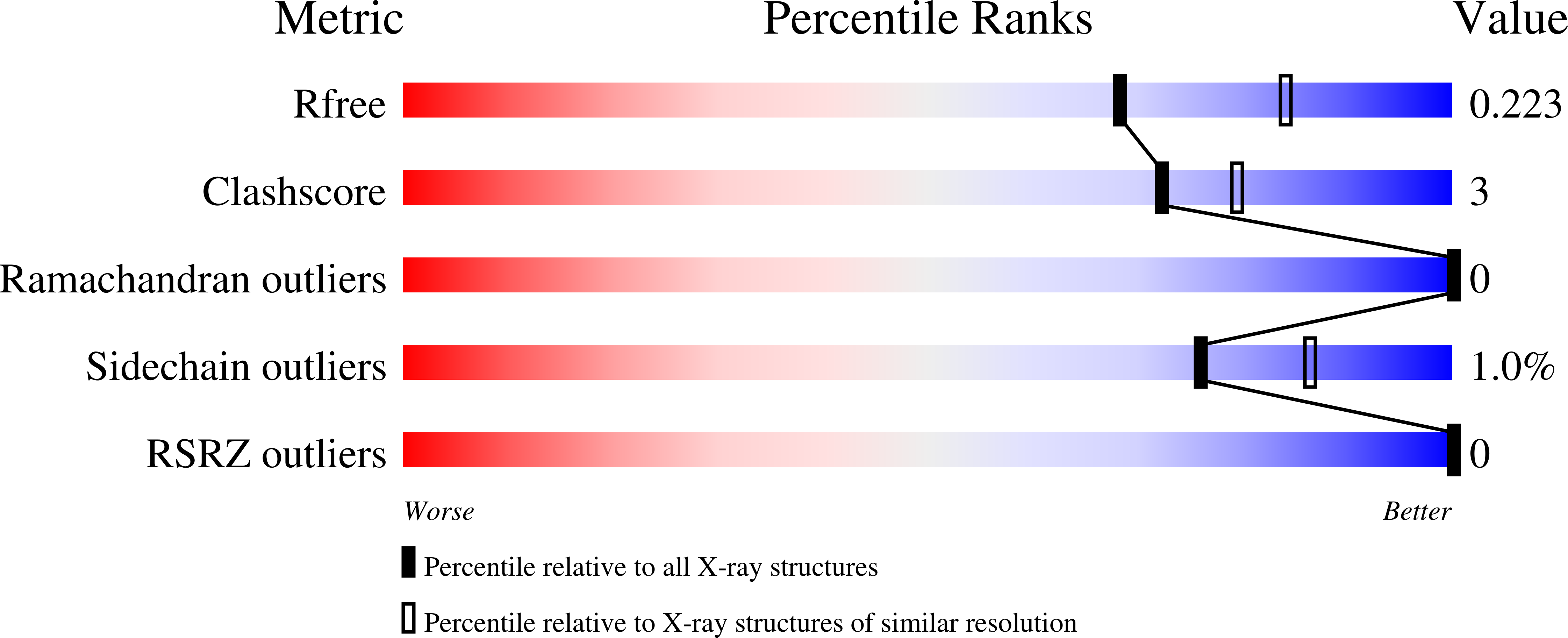
Identifying Oxacillinase-48 Carbapenemase Inhibitors Using DNA-Encoded Chemical Libraries.
Taylor, D.M., Anglin, J., Park, S., Ucisik, M.N., Faver, J.C., Simmons, N., Jin, Z., Palaniappan, M., Nyshadham, P., Li, F., Campbell, J., Hu, L., Sankaran, B., Prasad, B.V.V., Huang, H., Matzuk, M.M., Palzkill, T.(2020) ACS Infect Dis 6: 1214-1227
- PubMed: 32182432
- DOI: https://doi.org/10.1021/acsinfecdis.0c00015
- Primary Citation of Related Structures:
6UVK - PubMed Abstract:
Bacterial resistance to β-lactam antibiotics is largely mediated by β-lactamases, which catalyze the hydrolysis of these drugs and continue to emerge in response to antibiotic use. β-Lactamases that hydrolyze the last resort carbapenem class of β-lactam antibiotics (carbapenemases) are a growing global health threat. Inhibitors have been developed to prevent β-lactamase-mediated hydrolysis and restore the efficacy of these antibiotics. However, there are few inhibitors available for problematic carbapenemases such as oxacillinase-48 (OXA-48). A DNA-encoded chemical library approach was used to rapidly screen for compounds that bind and potentially inhibit OXA-48. Using this approach, a hit compound, CDD-97, was identified with submicromolar potency ( K i = 0.53 ± 0.08 μM) against OXA-48. X-ray crystallography showed that CDD-97 binds noncovalently in the active site of OXA-48. Synthesis and testing of derivatives of CDD-97 revealed structure-activity relationships and informed the design of a compound with a 2-fold increase in potency. CDD-97, however, synergizes poorly with β-lactam antibiotics to inhibit the growth of bacteria expressing OXA-48 due to poor accumulation into E. coli . Despite the low in vivo activity, CDD-97 provides new insights into OXA-48 inhibition and demonstrates the potential of using DNA-encoded chemistry technology to rapidly identify β-lactamase binders and to study β-lactamase inhibition, leading to clinically useful inhibitors.
Organizational Affiliation:
Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, Texas 77030, United States.