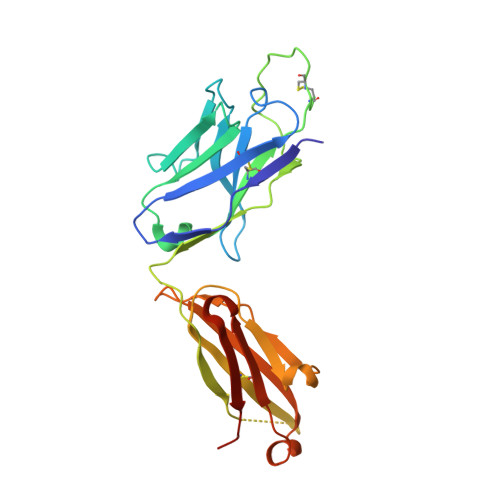
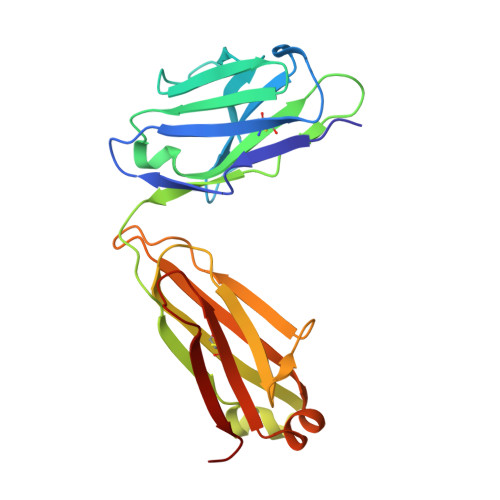
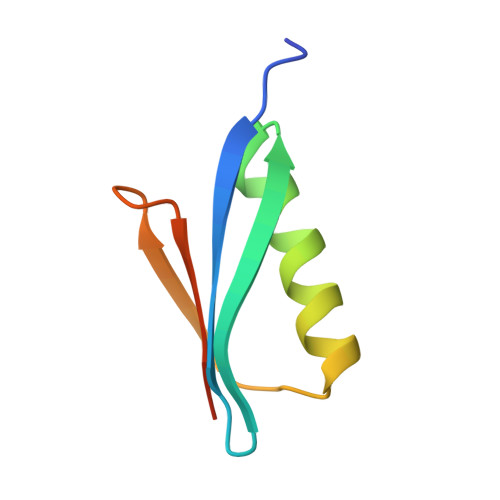
A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Kumar, S., Ju, B., Shapero, B., Lin, X., Ren, L., Zhang, L., Li, D., Zhou, Z., Feng, Y., Sou, C., Mann, C.J., Hao, Y., Sarkar, A., Hou, J., Nunnally, C., Hong, K., Wang, S., Ge, X., Su, B., Landais, E., Sok, D., Zwick, M.B., He, L., Zhu, J., Wilson, I.A., Shao, Y.(2020) Sci Adv 6
- PubMed: 32938661
- DOI: https://doi.org/10.1126/sciadv.abb1328
- Primary Citation of Related Structures:
6UUH, 6UUL, 6UUM - PubMed Abstract:
An oligomannose patch around the V3 base of HIV-1 envelope glycoprotein (Env) is recognized by multiple classes of broadly neutralizing antibodies (bNAbs). Here, we investigated the bNAb response to the V3 glycan supersite in an HIV-1-infected Chinese donor by Env-specific single B cell sorting, structural and functional studies, and longitudinal analysis of antibody and virus repertoires. Monoclonal antibodies 438-B11 and 438-D5 were isolated that potently neutralize HIV-1 with moderate breadth, are encoded by the V H 1-69 germline gene, and have a disulfide-linked long HCDR3 loop. Crystal structures of Env-bound and unbound antibodies revealed heavy chain-mediated recognition of the glycan supersite with a unique angle of approach and a critical role of the intra-HCDR3 disulfide. The mechanism of viral escape was examined via single-genome amplification/sequencing and glycan mutations around the N332 supersite. Our findings further emphasize the V3 glycan supersite as a prominent target for Env-based vaccine design.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.