Discovery and Structure-Based Optimization of Potent and Selective WD Repeat Domain 5 (WDR5) Inhibitors Containing a Dihydroisoquinolinone Bicyclic Core.
Tian, J., Teuscher, K.B., Aho, E.R., Alvarado, J.R., Mills, J.J., Meyers, K.M., Gogliotti, R.D., Han, C., Macdonald, J.D., Sai, J., Shaw, J.G., Sensintaffar, J.L., Zhao, B., Rietz, T.A., Thomas, L.R., Payne, W.G., Moore, W.J., Stott, G.M., Kondo, J., Inoue, M., Coffey, R.J., Tansey, W.P., Stauffer, S.R., Lee, T., Fesik, S.W.(2020) J Med Chem 63: 656-675
- PubMed: 31858797
- DOI: https://doi.org/10.1021/acs.jmedchem.9b01608
- Primary Citation of Related Structures:
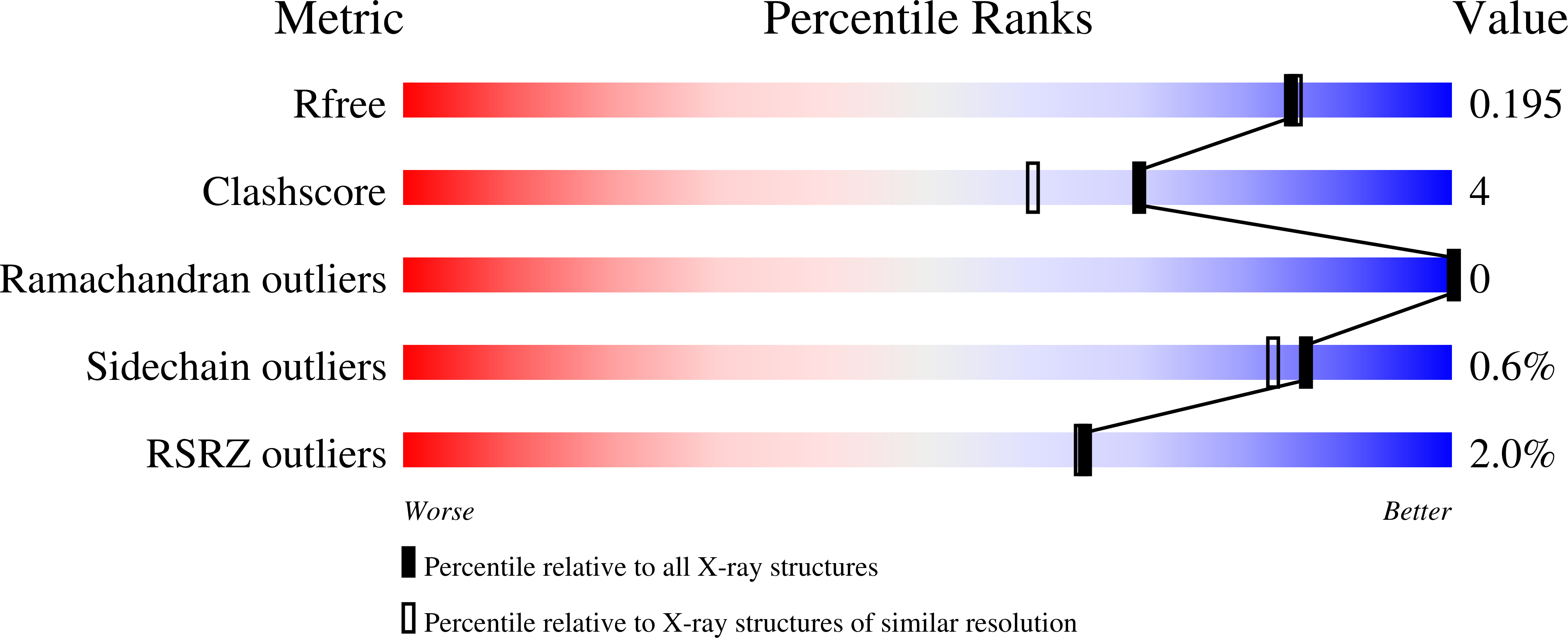
6UCS, 6UFX - PubMed Abstract:
WD repeat domain 5 (WDR5) is a member of the WD40-repeat protein family that plays a critical role in multiple chromatin-centric processes. Overexpression of WDR5 correlates with a poor clinical outcome in many human cancers, and WDR5 itself has emerged as an attractive target for therapy. Most drug-discovery efforts center on the WIN site of WDR5 that is responsible for the recruitment of WDR5 to chromatin. Here, we describe discovery of a novel WDR5 WIN site antagonists containing a dihydroisoquinolinone bicyclic core using a structure-based design. These compounds exhibit picomolar binding affinity and selective concentration-dependent antiproliferative activities in sensitive MLL-fusion cell lines. Furthermore, these WDR5 WIN site binders inhibit proliferation in MYC-driven cancer cells and reduce MYC recruitment to chromatin at MYC/WDR5 co-bound genes. Thus, these molecules are useful probes to study the implication of WDR5 inhibition in cancers and serve as a potential starting point toward the discovery of anti-WDR5 therapeutics.
Organizational Affiliation:
Leidos Biomedical Research , Frederick National Laboratory for Cancer Research , Frederick , Maryland 21701 , United States.