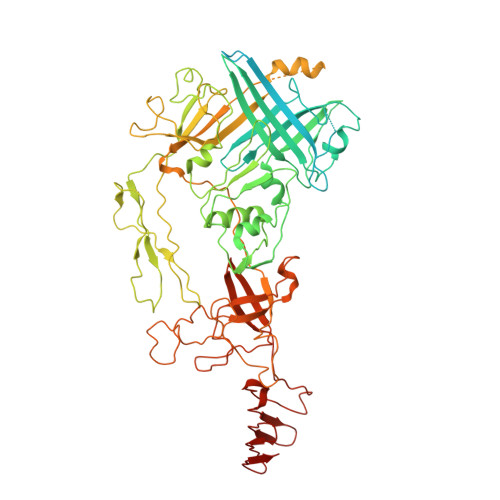
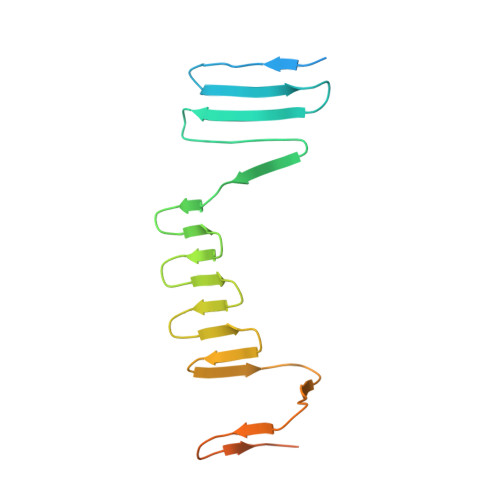Atomic Structure of the Francisella T6SS Central Spike Reveals a Unique alpha-Helical Lid and a Putative Cargo.
Yang, X., Clemens, D.L., Lee, B.Y., Cui, Y., Zhou, Z.H., Horwitz, M.A.(2019) Structure 27: 1811-1819.e6
- PubMed: 31677891
- DOI: https://doi.org/10.1016/j.str.2019.10.007
- Primary Citation of Related Structures:
6U9E, 6U9F, 6U9G - PubMed Abstract:
Francisella bacteria rely on a phylogenetically distinct type VI secretion system (T6SS) to escape host phagosomes and cause the fatal disease tularemia, but the structural and molecular mechanisms involved are unknown. Here we report the atomic structure of the Francisella T6SS central spike complex, obtained by cryo-electron microscopy. Our structural and functional studies demonstrate that, unlike the single-protein spike composition of other T6SS subtypes, Francisella T6SS's central spike is formed by two proteins, PdpA and VgrG, akin to T4-bacteriophage gp27 and gp5, respectively, and that PdpA has unique characteristics, including a putative cargo within its cavity and an N-terminal helical lid. Structure-guided mutagenesis demonstrates that the PdpA N-terminal lid and C-terminal spike are essential to Francisella T6SS function. PdpA is thus both an adaptor, connecting VgrG to the tube, and a likely carrier of secreted cargo. These findings are important to understanding Francisella pathogenicity and designing therapeutics to combat tularemia.
Organizational Affiliation:
Department of Microbiology, Immunology and Molecular Genetics, University of California, Los Angeles (UCLA), Los Angeles, CA 90095, USA; The California NanoSystems Institute (CNSI), University of California, Los Angeles (UCLA), Los Angeles, CA 90095, USA; State Key Laboratory of Medicinal Chemical Biology, Nankai University, 94 Weijin Road, Tianjin 300071, China.