Site-Specific Lysine Arylation as an Alternative Bioconjugation Strategy for Chemically Programmed Antibodies and Antibody-Drug Conjugates.
Hwang, D., Tsuji, K., Park, H., Burke Jr., T.R., Rader, C.(2019) Bioconjug Chem 30: 2889-2896
- PubMed: 31675216
- DOI: https://doi.org/10.1021/acs.bioconjchem.9b00609
- Primary Citation of Related Structures:
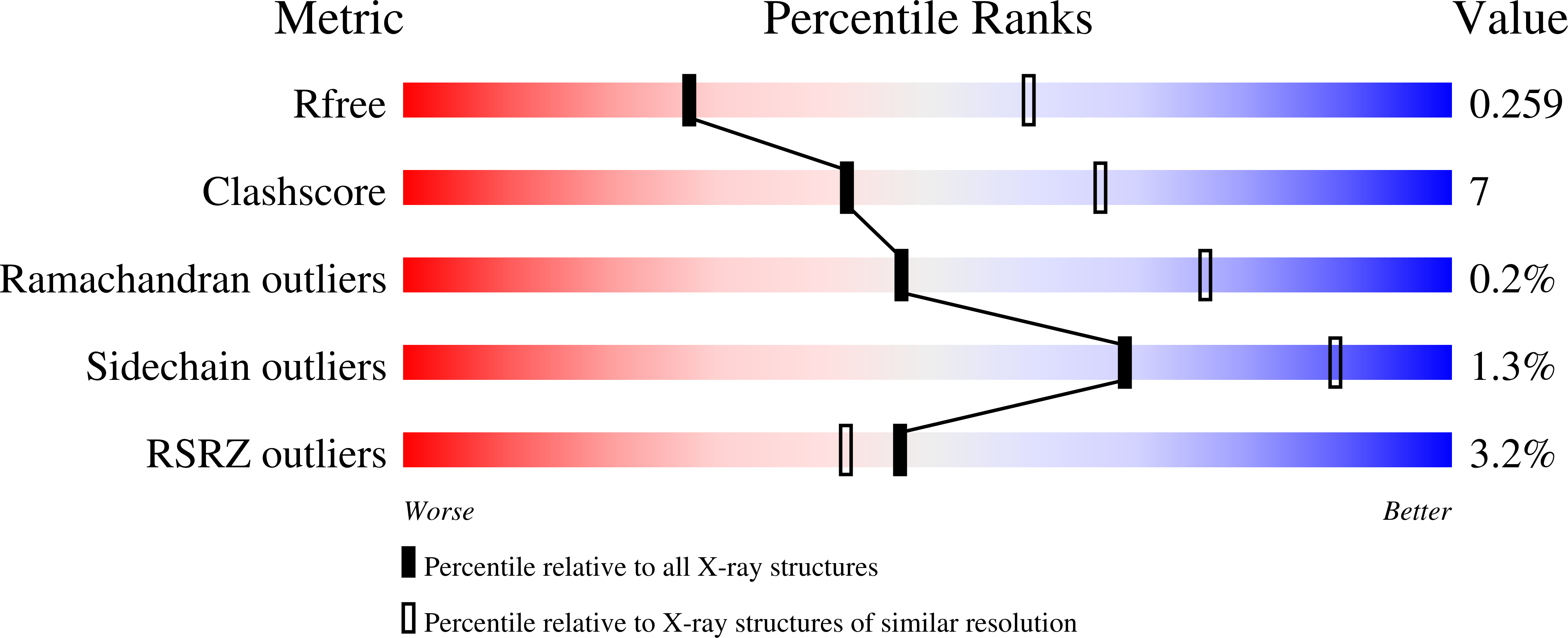
6U85 - PubMed Abstract:
By exploiting a uniquely reactive lysine residue (Lys99) for site-specific attachment of small molecules, the humanized catalytic antibody h38C2 has been used as bioconjugation module in the assembly of chemically programmed antibodies and antibody-drug conjugates. Treatment of h38C2 with β-lactam-functionalized small molecules has been previously shown to result in covalent conjugation by selective formation of a stable amide bond with the ε-amino group of the Lys99 residue. Here we report that heteroaryl methylsulfonyl (MS-PODA)-functionalized small molecules represent an alternative bioconjugation strategy through highly efficient, site-specific, and stable arylation of the Lys99 residue. A set of chemically programmed antibodies and antibody-drug conjugates assembled by Lys99 arylation provided proof-of-concept for the therapeutic utility of this alternative bioconjugation strategy. While being equally effective as β-lactam-functionalized ligands for bioconjugation with catalytic antibody h38C2, the MS-PODA moiety offers distinct synthetic advantages, making it highly attractive.
Organizational Affiliation:
Chemical Biology Laboratory, Center for Cancer Research , National Cancer Institute, National Institutes of Health , Frederick , Maryland 21702 , United States.