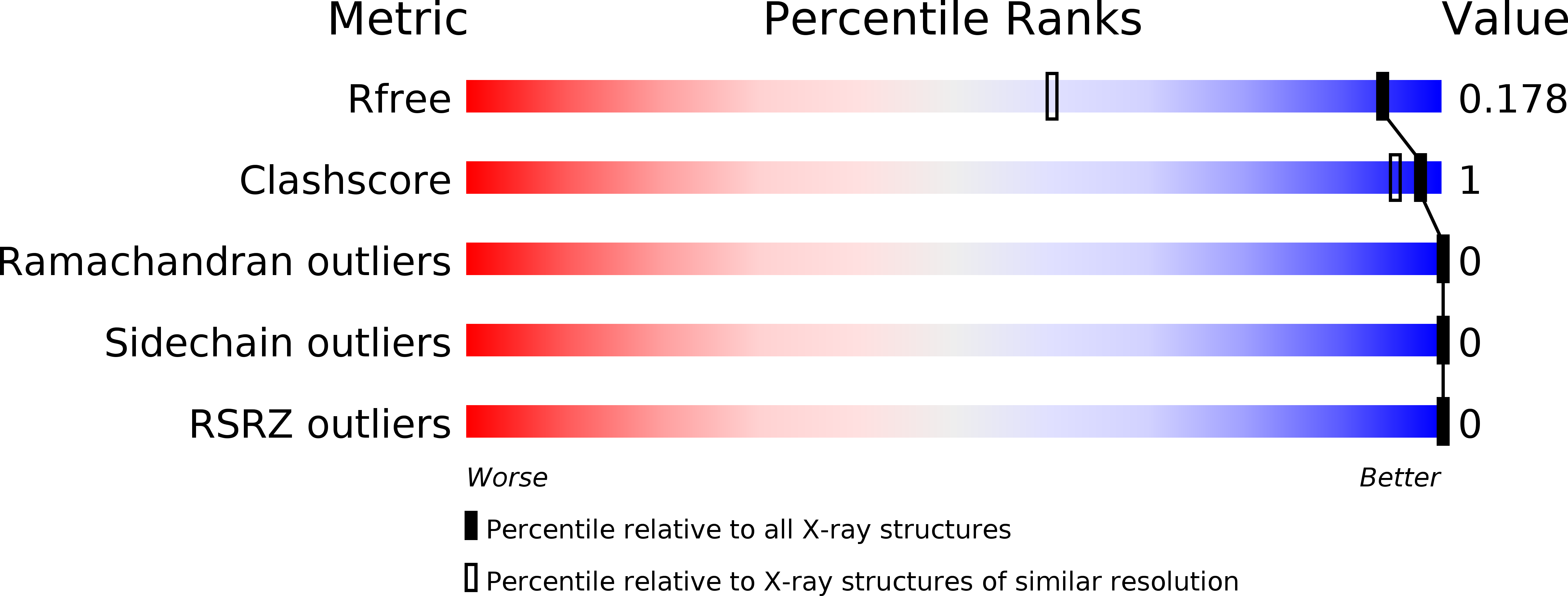
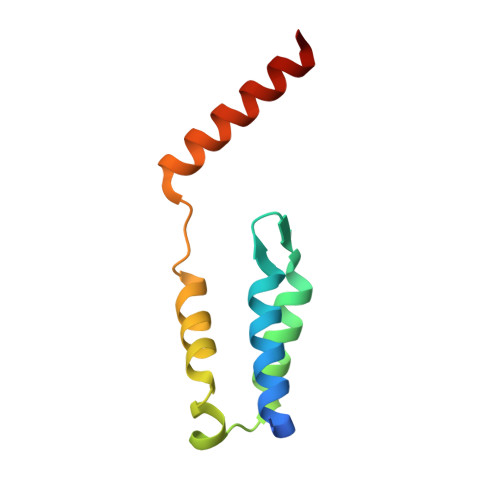
Fine-tuning of the LEDGF/p75 interaction network by dimerization
Lux, V., Brouns, T., Cermakova, K., Srb, P., Fabry, M., Madlikova, M., Horejsi, M., Kugler, M., Brynda, J., Novak, P., DeRijck, J., Christ, F., Debyser, Z., Veverka, V.(null) Structure