What does fluorine do to a protein? Thermodynamic, and highly-resolved structural insights into fluorine-labelled variants of the cold shock protein.
Welte, H., Zhou, T., Mihajlenko, X., Mayans, O., Kovermann, M.(2020) Sci Rep 10: 2640-2640
- PubMed: 32060391
- DOI: https://doi.org/10.1038/s41598-020-59446-w
- Primary Citation of Related Structures:
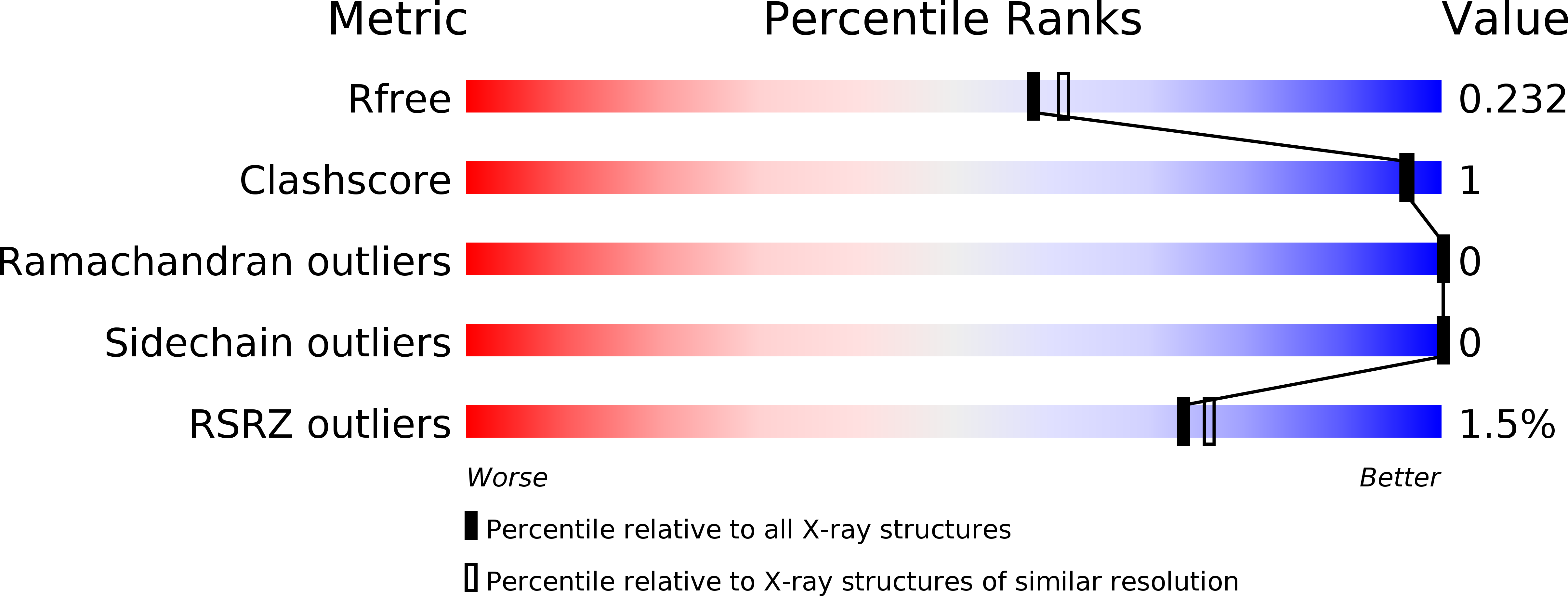
6SZZ, 6T00 - PubMed Abstract:
Fluorine labelling represents one promising approach to study proteins in their native environment due to efficient suppressing of background signals. Here, we systematically probe inherent thermodynamic and structural characteristics of the Cold shock protein B from Bacillus subtilis (BsCspB) upon fluorine labelling. A sophisticated combination of fluorescence and NMR experiments has been applied to elucidate potential perturbations due to insertion of fluorine into the protein. We show that single fluorine labelling of phenylalanine or tryptophan residues has neither significant impact on thermodynamic stability nor on folding kinetics compared to wild type BsCspB. Structure determination of fluorinated phenylalanine and tryptophan labelled BsCspB using X-ray crystallography reveals no displacements even for the orientation of fluorinated aromatic side chains in comparison to wild type BsCspB. Hence we propose that single fluorinated phenylalanine and tryptophan residues used for protein labelling may serve as ideal probes to reliably characterize inherent features of proteins that are present in a highly biological context like the cell.
Organizational Affiliation:
Department of Chemistry, Universitätsstrasse 10, Universität Konstanz, DE-78457, Konstanz, Germany.