Design, Synthesis, and Biological Evaluation of Novel Indoles Targeting the Influenza PB2 Cap Binding Region.
McGowan, D.C., Balemans, W., Embrechts, W., Motte, M., Keown, J.R., Buyck, C., Corbera, J., Funes, M., Moreno, L., Cooymans, L., Tahri, A., Eymard, J., Stoops, B., Strijbos, R., Van den Berg, J., Fodor, E., Grimes, J.M., Koul, A., Jonckers, T.H.M., Raboisson, P., Guillemont, J.(2019) J Med Chem 62: 9680-9690
- PubMed: 31647875
- DOI: https://doi.org/10.1021/acs.jmedchem.9b01091
- Primary Citation of Related Structures:
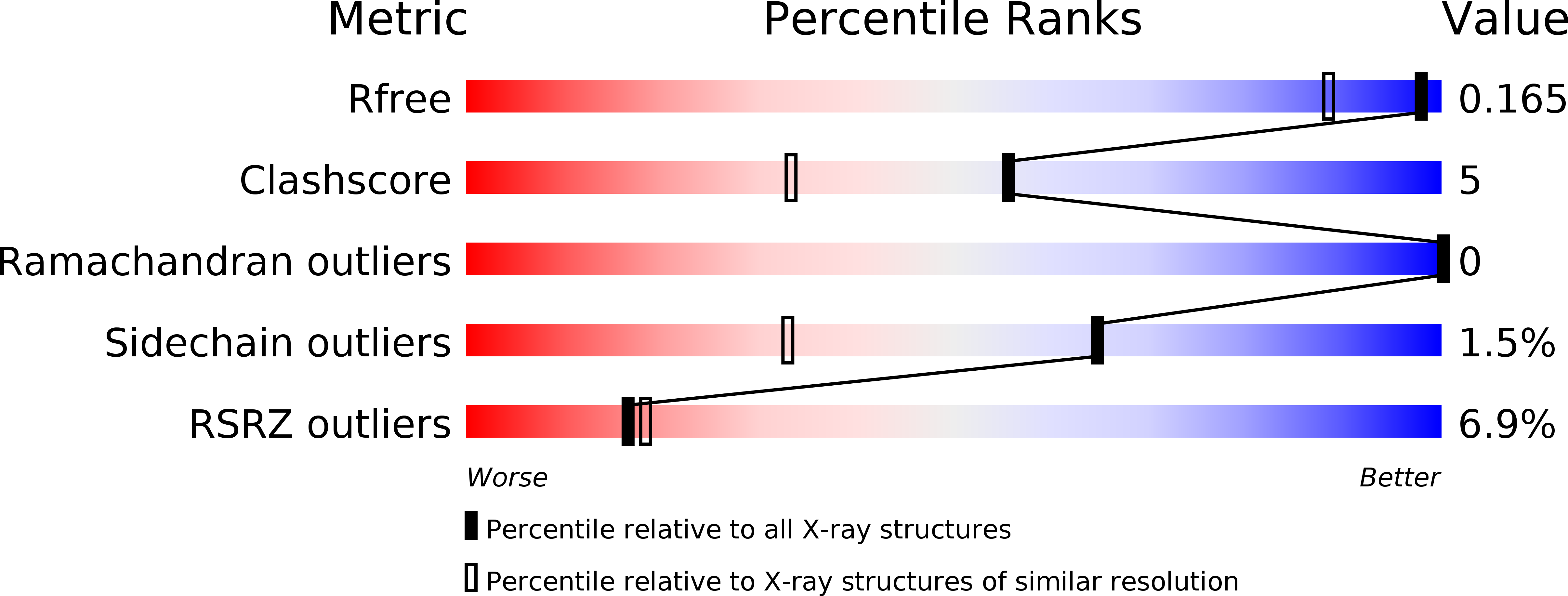
6S5V - PubMed Abstract:
In the search for novel influenza inhibitors we evaluated 7-fluoro-substituted indoles as bioisosteric replacements for the 7-azaindole scaffold of Pimodivir, a PB2 (polymerase basic protein 2) inhibitor currently in clinical development. Specifically, a 5,7-difluoroindole derivative 11a was identified as a potent and metabolically stable influenza inhibitor. 11a demonstrated a favorable oral pharmacokinetic profile and in vivo efficacy in mice. In addition, it was found that 11a was not at risk of metabolism via aldehyde oxidase, an advantage over previously described inhibitors of this class. The crystal structure of 11a bound to influenza A PB2 cap region is disclosed here and deposited to the PDB.
Organizational Affiliation:
Janssen Pharmaceutica, N.V. , Turnhoutseweg 30 , 2340 Beerse , Belgium.