X-Ray Crystal Structure of a Second Generation Peptide Dendrimer in Complex with Pseudomonas aeruginosa Lectin LecB
Baeriswyl, S., Javor, S., Stocker, A., Darbre, T., Reymond, J.-L.(2019) Helv Chim Acta
Experimental Data Snapshot
(2019) Helv Chim Acta
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fucose-binding lectin | 115 | Pseudomonas aeruginosa | Mutation(s): 0 Gene Names: lecB, C0044_25260, CAZ10_21840, DT376_00595, DY979_15445, ECC04_10105, EFK27_13700, EGV95_09240, EGY23_15550, IPC669_23070... |  | |
UniProt | |||||
Find proteins for Q9HYN5 (Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1)) Explore Q9HYN5 Go to UniProtKB: Q9HYN5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9HYN5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| SBD6 | E [auth F], F [auth H] | 3 | synthetic construct | Mutation(s): 0 | 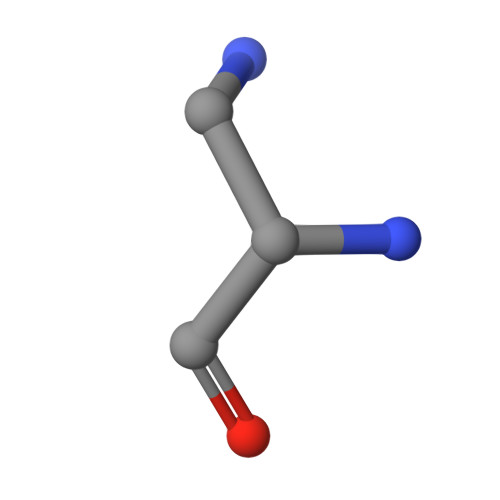 |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZDC (Subject of Investigation/LOI) Query on ZDC | I [auth A], K [auth A], O [auth C], R [auth H] | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid C8 H14 O6 YTZUDUWVQZSKNN-OASCRQMUSA-N |  | ||
| CA Query on CA | G [auth A] H [auth A] J [auth A] L [auth B] M [auth C] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| DPP Query on DPP | E [auth F], F [auth H] | L-PEPTIDE LINKING | C3 H8 N2 O2 |  | ALA |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 91.003 | α = 90 |
| b = 91.003 | β = 90 |
| c = 315.583 | γ = 120 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Swiss National Science Foundation | Switzerland | -- |