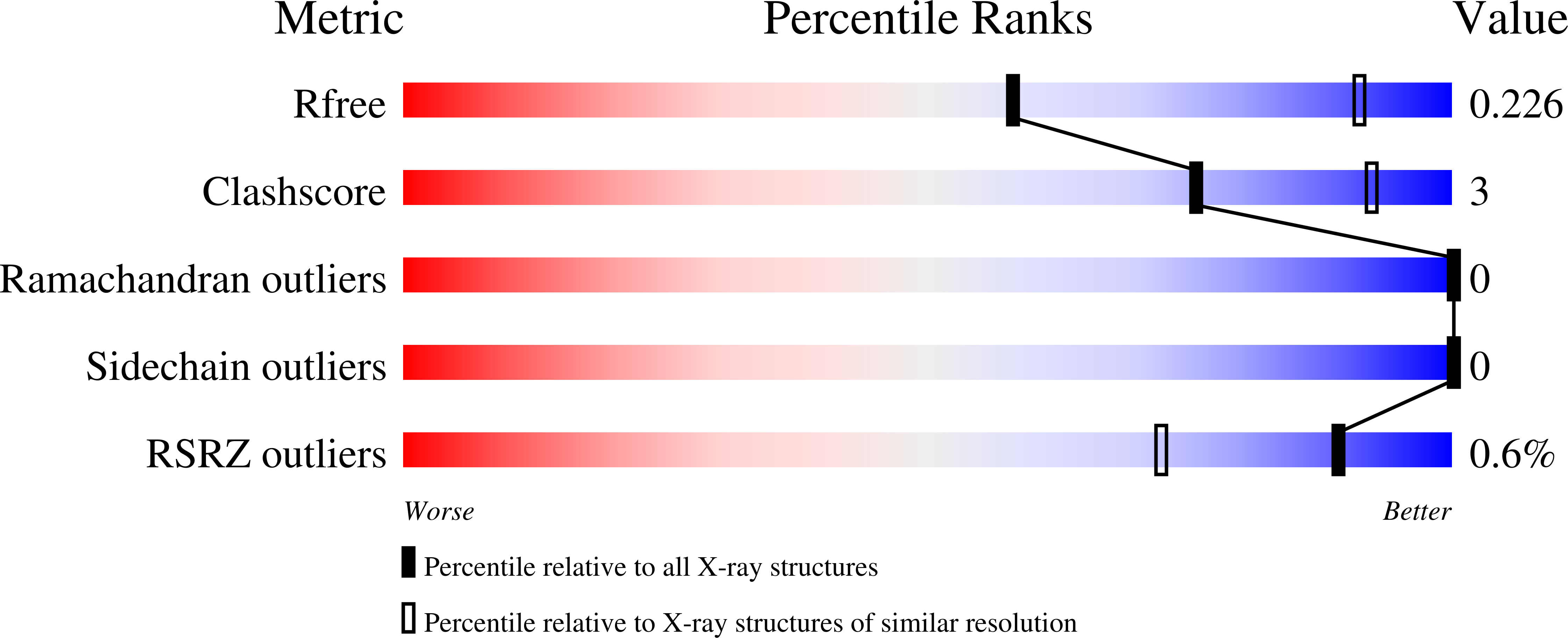
A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Mrugala, B., Milaczewska, A., Porebski, P.J., Niedzialkowska, E., Guzik, M., Minor, W., Borowski, T.(2021) FEBS J 288: 1366-1386
- PubMed: 32592631
- DOI: https://doi.org/10.1111/febs.15462
- Primary Citation of Related Structures:
6S0R, 6S0S, 6S0T, 6S0U, 6S0V, 6S0W - PubMed Abstract:
Kanamycin A is an aminoglycoside antibiotic isolated from Streptomyces kanamyceticus and used against a wide spectrum of bacteria, including Mycobacterium tuberculosis. Biosynthesis of kanamycin involves an oxidative deamination step catalyzed by kanamycin B dioxygenase (KanJ), thereby the C2' position of kanamycin B is transformed into a keto group upon release of ammonia. Here, we present for the first time, structural models of KanJ with several ligands, which along with the results of ITC binding assays and HPLC activity tests explain substrate specificity of the enzyme. The large size of the binding pocket suggests that KanJ can accept a broad range of substrates, which was confirmed by activity tests. Specificity of the enzyme with respect to its substrate is determined by the hydrogen bond interactions between the methylamino group of the antibiotic and highly conserved Asp134 and Cys150 as well as between hydroxyl groups of the substrate and Asn120 and Gln80. Upon antibiotic binding, the C terminus loop is significantly rearranged and Gln80 and Asn120, which are directly involved in substrate recognition, change their conformations. Based on reaction energy profiles obtained by density functional theory (DFT) simulations, we propose a mechanism of ketone formation involving the reactive Fe IV = O and proceeding either via OH rebound, which yields a hemiaminal intermediate or by abstraction of two hydrogen atoms, which leads to an imine species. At acidic pH, the latter involves a lower barrier than the OH rebound, whereas at basic pH, the barrier leading to an imine vanishes completely. DATABASES: Structural data are available in PDB database under the accession numbers: 6S0R, 6S0T, 6S0U, 6S0W, 6S0V, 6S0S. Diffraction images are available at the Integrated Resource for Reproducibility in Macromolecular Crystallography at http://proteindiffraction.org under DOIs: 10.18430/m36s0t, 10.18430/m36s0u, 10.18430/m36s0r, 10.18430/m36s0s, 10.18430/m36s0v, 10.18430/m36s0w. A data set collection of computational results is available in the Mendeley Data database under DOI: 10.17632/sbyzssjmp3.1 and in the ioChem-BD database under DOI: 10.19061/iochem-bd-4-18.
Organizational Affiliation:
Jerzy Haber Institute of Catalysis and Surface Chemistry, Polish Academy of Sciences, Krakow, Poland.