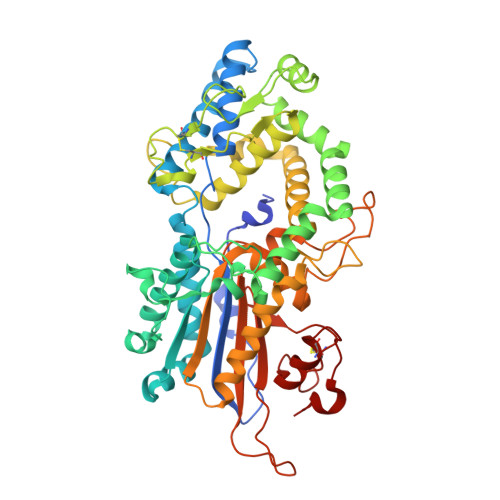
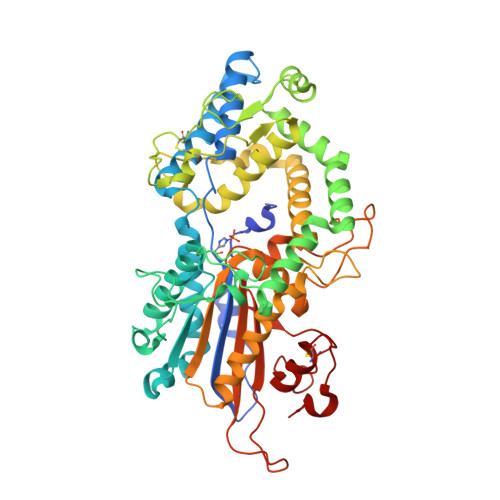
Snapshots during the catalytic cycle of a histidine acid phytase reveal an induced-fit structural mechanism.
Acquistapace, I.M., Zi Etek, M.A., Li, A.W.H., Salmon, M., Kuhn, I., Bedford, M.R., Brearley, C.A., Hemmings, A.M.(2020) J Biol Chem 295: 17724-17737
- PubMed: 33454010
- DOI: https://doi.org/10.1074/jbc.RA120.015925
- Primary Citation of Related Structures:
6RXD, 6RXE, 6RXF, 6RXG - PubMed Abstract:
Highly engineered phytases, which sequentially hydrolyze the hexakisphosphate ester of inositol known as phytic acid, are routinely added to the feeds of monogastric animals to improve phosphate bioavailability. New phytases are sought as starting points to further optimize the rate and extent of dephosphorylation of phytate in the animal digestive tract. Multiple inositol polyphosphate phosphatases (MINPPs) are clade 2 histidine phosphatases (HP2P) able to carry out the stepwise hydrolysis of phytate. MINPPs are not restricted by a strong positional specificity making them attractive targets for development as feed enzymes. Here, we describe the characterization of a MINPP from the Gram-positive bacterium Bifidobacterium longum (BlMINPP). BlMINPP has a typical HP2P-fold but, unusually, possesses a large α-domain polypeptide insertion relative to other MINPPs. This insertion, termed the U-loop, spans the active site and contributes to substrate specificity pockets underpopulated in other HP2Ps. Mutagenesis of U-loop residues reveals its contribution to enzyme kinetics and thermostability. Moreover, four crystal structures of the protein along the catalytic cycle capture, for the first time in an HP2P, a large ligand-driven α-domain motion essential to allow substrate access to the active site. This motion recruits residues both downstream of a molecular hinge and on the U-loop to participate in specificity subsites, and mutagenesis identified a mobile lysine residue as a key determinant of positional specificity of the enzyme. Taken together, these data provide important new insights to the factors determining stability, substrate recognition, and the structural mechanism of hydrolysis in this industrially important group of enzymes.
Organizational Affiliation:
School of Biological Sciences, University of East Anglia, Norwich, United Kingdom.