Structural basis for the interaction of the chaperone Cbp3 with newly synthesized cytochromebduring mitochondrial respiratory chain assembly.
Ndi, M., Masuyer, G., Dawitz, H., Carlstrom, A., Michel, M., Elofsson, A., Rapp, M., Stenmark, P., Ott, M.(2019) J Biol Chem 294: 16663-16671
- PubMed: 31537648
- DOI: https://doi.org/10.1074/jbc.RA119.010483
- Primary Citation of Related Structures:
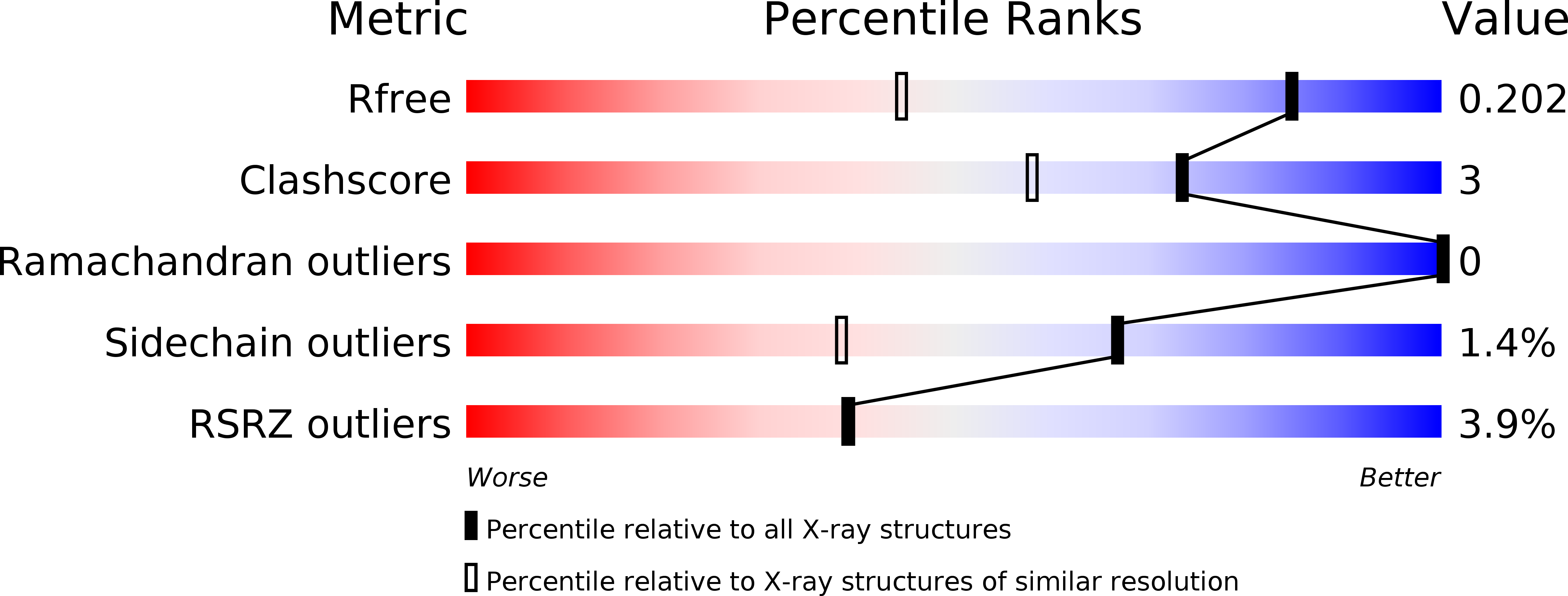
6RWT - PubMed Abstract:
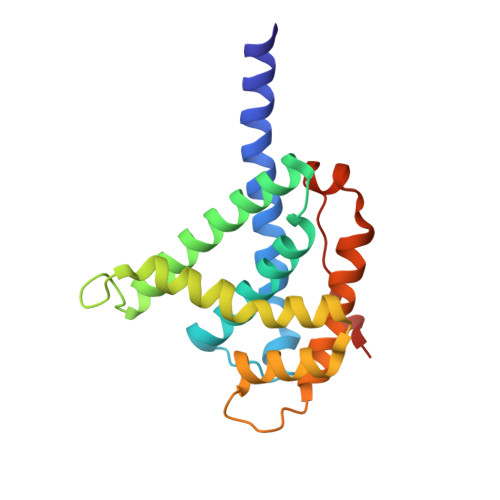
Assembly of the mitochondrial respiratory chain requires the coordinated synthesis of mitochondrial and nuclear encoded subunits, redox co-factor acquisition, and correct joining of the subunits to form functional complexes. The conserved Cbp3-Cbp6 chaperone complex binds newly synthesized cytochrome b and supports the ordered acquisition of the heme co-factors. Moreover, it functions as a translational activator by interacting with the mitoribosome. Cbp3 consists of two distinct domains: an N-terminal domain present in mitochondrial Cbp3 homologs and a highly conserved C-terminal domain comprising a ubiquinol-cytochrome c chaperone region. Here, we solved the crystal structure of this C-terminal domain from a bacterial homolog at 1.4 Å resolution, revealing a unique all-helical fold. This structure allowed mapping of the interaction sites of yeast Cbp3 with Cbp6 and cytochrome b via site-specific photo-cross-linking. We propose that mitochondrial Cbp3 homologs carry an N-terminal extension that positions the conserved C-terminal domain at the ribosomal tunnel exit for an efficient interaction with its substrate, the newly synthesized cytochrome b protein.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Stockholm University SE-10691 Stockholm, Sweden.