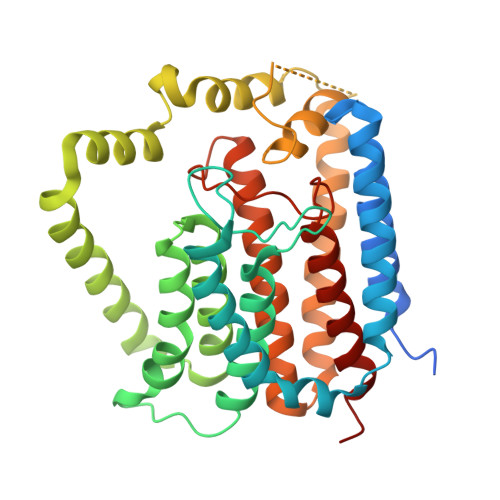
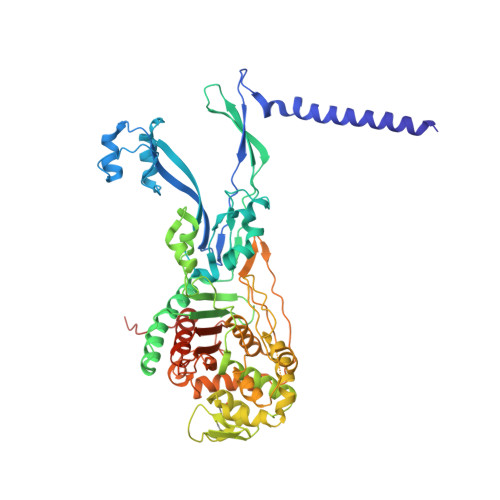
Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Sjodt, M., Rohs, P.D.A., Gilman, M.S.A., Erlandson, S.C., Zheng, S., Green, A.G., Brock, K.P., Taguchi, A., Kahne, D., Walker, S., Marks, D.S., Rudner, D.Z., Bernhardt, T.G., Kruse, A.C.(2020) Nat Microbiol 5: 813-820
- PubMed: 32152588
- DOI: https://doi.org/10.1038/s41564-020-0687-z
- Primary Citation of Related Structures:
6PL5, 6PL6 - PubMed Abstract:
The shape, elongation, division and sporulation (SEDS) proteins are a highly conserved family of transmembrane glycosyltransferases that work in concert with class B penicillin-binding proteins (bPBPs) to build the bacterial peptidoglycan cell wall 1-6 . How these proteins coordinate polymerization of new glycan strands with their crosslinking to the existing peptidoglycan meshwork is unclear. Here, we report the crystal structure of the prototypical SEDS protein RodA from Thermus thermophilus in complex with its cognate bPBP at 3.3 Å resolution. The structure reveals a 1:1 stoichiometric complex with two extensive interaction interfaces between the proteins: one in the membrane plane and the other at the extracytoplasmic surface. When in complex with a bPBP, RodA shows an approximately 10 Å shift of transmembrane helix 7 that exposes a large membrane-accessible cavity. Negative-stain electron microscopy reveals that the complex can adopt a variety of different conformations. These data define the bPBP pedestal domain as the key allosteric activator of RodA both in vitro and in vivo, explaining how a SEDS-bPBP complex can coordinate its dual enzymatic activities of peptidoglycan polymerization and crosslinking to build the cell wall.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Blavatnik Institute, Harvard Medical School, Boston, MA, USA.