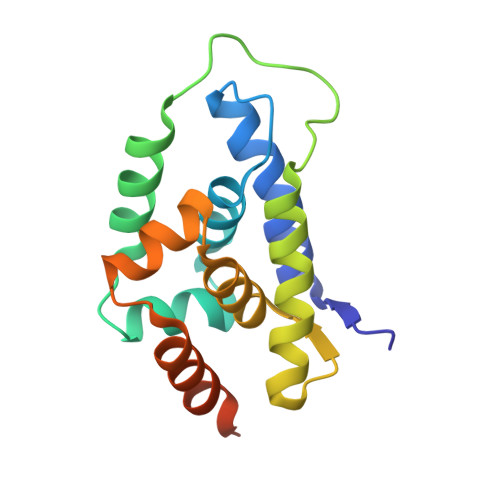
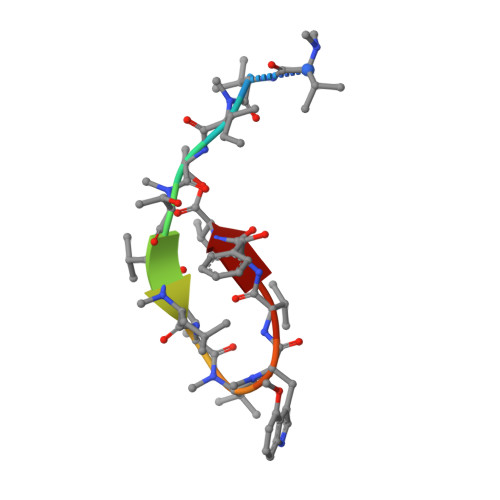
Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Wolf, N.M., Lee, H., Zagal, D., Nam, J.W., Oh, D.C., Lee, H., Suh, J.W., Pauli, G.F., Cho, S., Abad-Zapatero, C.(2020) Acta Crystallogr D Struct Biol 76: 458-471
- PubMed: 32355042
- DOI: https://doi.org/10.1107/S2059798320004027
- Primary Citation of Related Structures:
6PBA, 6PBQ, 6PBS, 6UCR - PubMed Abstract:
The biological processes related to protein homeostasis in Mycobacterium tuberculosis, the etiologic agent of tuberculosis, have recently been established as critical pathways for therapeutic intervention. Proteins of particular interest are ClpC1 and the ClpC1-ClpP1-ClpP2 proteasome complex. The structure of the potent antituberculosis macrocyclic depsipeptide ecumicin complexed with the N-terminal domain of ClpC1 (ClpC1-NTD) is presented here. Crystals of the ClpC1-NTD-ecumicin complex were monoclinic (unit-cell parameters a = 80.0, b = 130.0, c = 112.0 Å, β = 90.07°; space group P2 1 ; 12 complexes per asymmetric unit) and diffracted to 2.5 Å resolution. The structure was solved by molecular replacement using the self-rotation function to resolve space-group ambiguities. The new structure of the ecumicin complex showed a unique 1:2 (target:ligand) stoichiometry exploiting the intramolecular dyad in the α-helical fold of the target N-terminal domain. The structure of the ecumicin complex unveiled extensive interactions in the uniquely extended N-terminus, a critical binding site for the known cyclopeptide complexes. This structure, in comparison with the previously reported rufomycin I complex, revealed unique features that could be relevant for understanding the mechanism of action of these potential antituberculosis drug leads. Comparison of the ecumicin complex and the ClpC1-NTD-L92S/L96P double-mutant structure with the available structures of rufomycin I and cyclomarin A complexes revealed a range of conformational changes available to this small N-terminal helical domain and the minor helical alterations involved in the antibiotic-resistance mechanism. The different modes of binding and structural alterations could be related to distinct modes of action.
Organizational Affiliation:
Institute for Tuberculosis Research, College of Pharmacy, University of Illinois at Chicago, Chicago, IL 60612, USA.