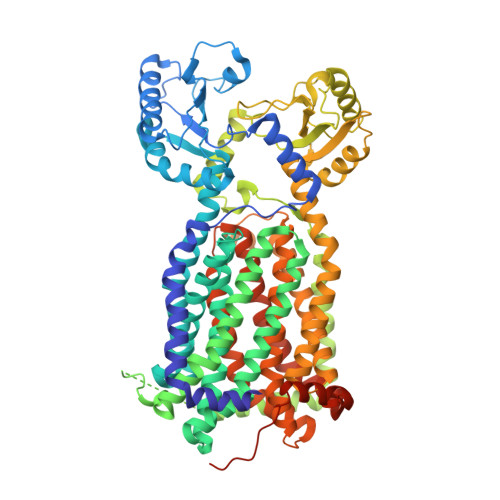
MmpL3 is a lipid transporter that binds trehalose monomycolate and phosphatidylethanolamine.
Su, C.C., Klenotic, P.A., Bolla, J.R., Purdy, G.E., Robinson, C.V., Yu, E.W.(2019) Proc Natl Acad Sci U S A 116: 11241-11246
- PubMed: 31113875
- DOI: https://doi.org/10.1073/pnas.1901346116
- Primary Citation of Related Structures:
6OR2 - PubMed Abstract:
The cell envelope of Mycobacterium tuberculosis is notable for the abundance of mycolic acids (MAs), essential to mycobacterial viability, and of other species-specific lipids. The mycobacterial cell envelope is extremely hydrophobic, which contributes to virulence and antibiotic resistance. However, exactly how fatty acids and lipidic elements are transported across the cell envelope for cell-wall biosynthesis is unclear. Mycobacterial membrane protein Large 3 (MmpL3) is essential and required for transport of trehalose monomycolates (TMMs), precursors of MA-containing trehalose dimycolates (TDM) and mycolyl arabinogalactan peptidoglycan, but the exact function of MmpL3 remains elusive. Here, we report a crystal structure of Mycobacterium smegmatis MmpL3 at a resolution of 2.59 Å, revealing a monomeric molecule that is structurally distinct from all known bacterial membrane proteins. A previously unknown MmpL3 ligand, phosphatidylethanolamine (PE), was discovered inside this transporter. We also show, via native mass spectrometry, that MmpL3 specifically binds both TMM and PE, but not TDM, in the micromolar range. These observations provide insight into the function of MmpL3 and suggest a possible role for this protein in shuttling a variety of lipids to strengthen the mycobacterial cell wall.
Organizational Affiliation:
Department of Pharmacology, Case Western Reserve University School of Medicine, Cleveland, OH 44106.