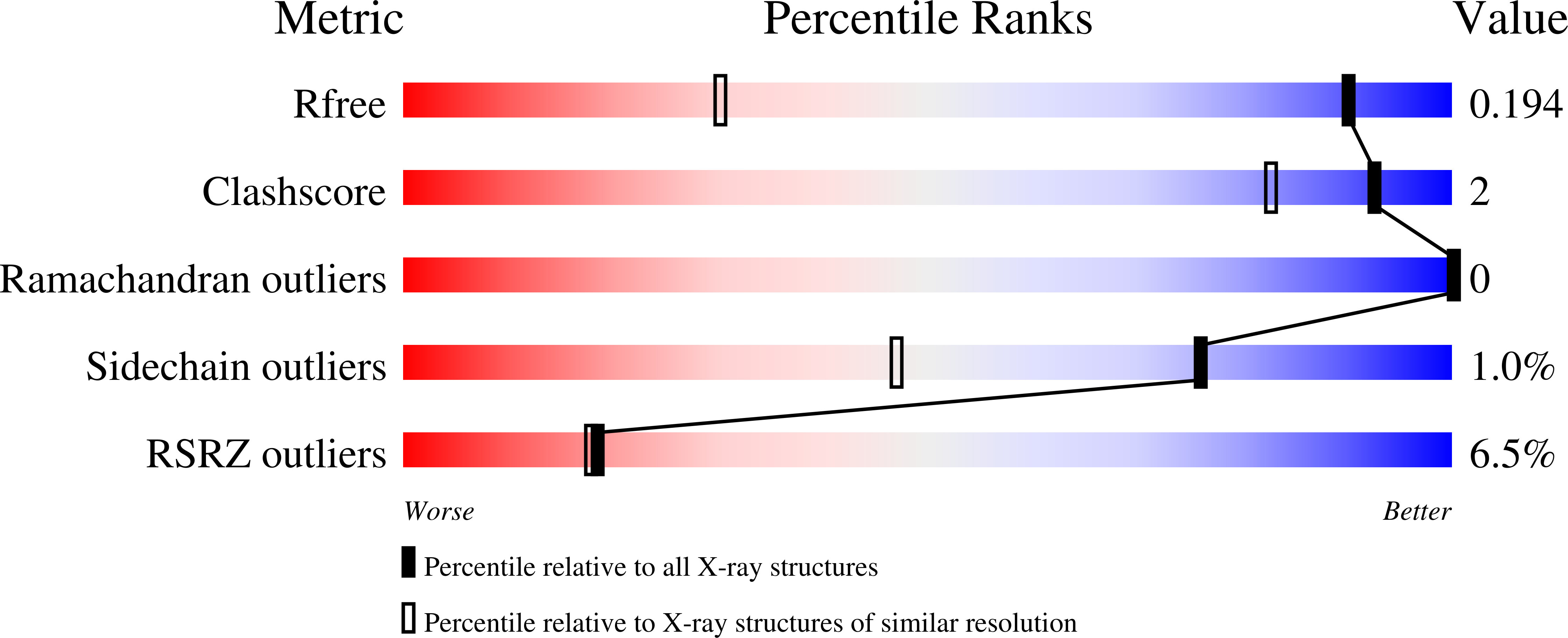
Importance of the beta 5-beta 6 Loop for the Structure, Catalytic Efficiency, and Stability of Carbapenem-Hydrolyzing Class D beta-Lactamase Subfamily OXA-143.
Antunes, V.U., Llontop, E.E., Vasconcelos, F.N.D.C., Lopez de Los Santos, Y., Oliveira, R.J., Lincopan, N., Farah, C.S., Doucet, N., Mittermaier, A., Favaro, D.C.(2019) Biochemistry 58: 3604-3616
- PubMed: 31355630
- DOI: https://doi.org/10.1021/acs.biochem.9b00365
- Primary Citation of Related Structures:
6NZ8 - PubMed Abstract:
The class D β-lactamase OXA-143 has been described as an efficient penicillinase, oxacillinase, and carbapenemase. The D224A variant, known as OXA-231, was described in 2012 as exhibiting less activity toward imipenem and increased oxacillinase activity. Additionally, the P227S mutation was reported as a case of convergent evolution for homologous enzymes. To investigate the impact of both mutations (D224A and P227S), we describe in this paper a deep investigation of the enzymatic activities of these three homologues. OXA-143(P227S) presented enhanced catalytic activity against ampicillin, oxacillins, aztreonam, and carbapenems. In addition, OXA-143(P227S) was the only member capable of hydrolyzing ceftazidime. These enhanced activities were due to a combination of a higher affinity (lower K m ) and a higher turnover number (higher k cat ). We also determined the crystal structure of apo OXA-231. As expected, the structure of this variant is very similar to the published OXA-143 structure, except for the two M223 conformations and the absence of electron density for three solvent-exposed loop segments. Molecular dynamics calculations showed that both mutants experience higher flexibility compared to that of the wild-type form. Therefore, our results illustrate that D224A and P227S act as deleterious and positive mutations, respectively, within the evolutionary path of the OXA-143 subfamily toward a more efficient carbapenemase.
Organizational Affiliation:
Department of Organic Chemistry , State University of Campinas , São Paulo , SP 13083-970 , Brazil.