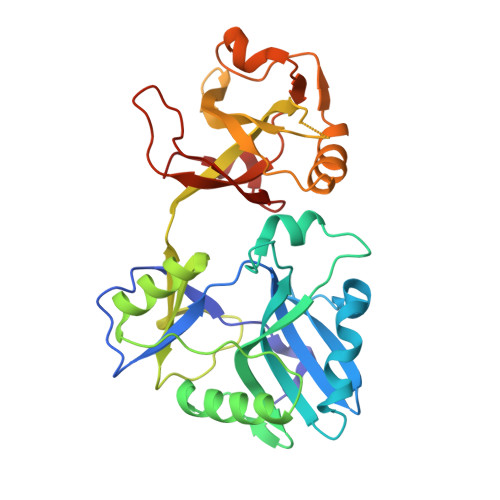
Structures of ATP-bound DNA ligase D in a closed domain conformation reveal a network of amino acid and metal contacts to the ATP phosphates.
Unciuleac, M.C., Goldgur, Y., Shuman, S.(2019) J Biol Chem 294: 5094-5104
- PubMed: 30718283
- DOI: https://doi.org/10.1074/jbc.RA119.007445
- Primary Citation of Related Structures:
6NHX, 6NHZ - PubMed Abstract:
DNA ligases are the sine qua non of genome integrity and essential for DNA replication and repair in all organisms. DNA ligases join 3'-OH and 5'-PO 4 ends via a series of three nucleotidyl transfer steps. In step 1, ligase reacts with ATP or NAD + to form a covalent ligase-(lysyl-Nζ)-AMP intermediate and release pyrophosphate (PP i ) or nicotinamide mononucleotide. In step 2, AMP is transferred from ligase-adenylate to the 5'-PO 4 DNA end to form a DNA-adenylate intermediate (AppDNA). In step 3, ligase catalyzes attack by a DNA 3'-OH on the DNA-adenylate to seal the two ends via a phosphodiester bond and release AMP. Eukaryal, archaeal, and many bacterial and viral DNA ligases are ATP-dependent. The catalytic core of ATP-dependent DNA ligases consists of an N-terminal nucleotidyltransferase domain fused to a C-terminal OB domain. Here we report crystal structures at 1.4-1.8 Å resolution of Mycobacterium tuberculosis LigD, an ATP-dependent DNA ligase dedicated to nonhomologous end joining, in complexes with ATP that highlight large movements of the OB domain (∼50 Å), from a closed conformation in the ATP complex to an open conformation in the covalent ligase-AMP intermediate. The LigD·ATP structures revealed a network of amino acid contacts to the ATP phosphates that stabilize the transition state and orient the PP i leaving group. A complex with ATP and magnesium suggested a two-metal mechanism of lysine adenylylation driven by a catalytic Mg 2+ that engages the ATP α phosphate and a second metal that bridges the ATP β and γ phosphates.
Organizational Affiliation:
From the Molecular Biology and.