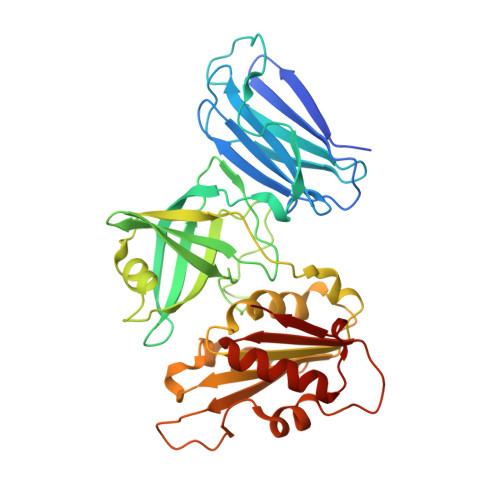
Crystal structures of the naturally fused CS and cytochrome b5reductase (b5R) domains of Ncb5or reveal an expanded CS fold, extensive CS-b5R interactions and productive binding of the NAD(P)+nicotinamide ring.
Benson, D.R., Lovell, S., Mehzabeen, N., Galeva, N., Cooper, A., Gao, P., Battaile, K.P., Zhu, H.(2019) Acta Crystallogr D Struct Biol 75: 628-638
- PubMed: 31282472
- DOI: https://doi.org/10.1107/S205979831900754X
- Primary Citation of Related Structures:
6MV1, 6MV2 - PubMed Abstract:
Ncb5or (NADH-cytochrome b 5 oxidoreductase), a cytosolic ferric reductase implicated in diabetes and neurological diseases, comprises three distinct domains, cytochrome b 5 (b 5 ) and cytochrome b 5 reductase (b 5 R) domains separated by a CHORD-Sgt1 (CS) domain, and a novel 50-residue N-terminal region. Understanding how interdomain interactions in Ncb5or facilitate the shuttling of electrons from NAD(P)H to heme, and how the process compares with the microsomal b 5 (Cyb5A) and b 5 R (Cyb5R3) system, is of interest. A high-resolution structure of the b 5 domain (PDB entry 3lf5) has previously been reported, which exhibits substantial differences in comparison to Cyb5A. The structural characterization of a construct comprising the naturally fused CS and b 5 R domains with bound FAD and NAD + (PDB entry 6mv1) or NADP + (PDB entry 6mv2) is now reported. The structures reveal that the linker between the CS and b 5 R cores is more ordered than predicted, with much of it extending the β-sandwich motif of the CS domain. This limits the flexibility between the two domains, which recognize one another via a short β-sheet motif and a network of conserved side-chain hydrogen bonds, salt bridges and cation-π interactions. Notable differences in FAD-protein interactions in Ncb5or and Cyb5R3 provide insight into the selectivity for docking of their respective b 5 redox partners. The structures also afford a structural explanation for the unusual ability of Ncb5or to utilize both NADH and NADPH, and represent the first examples of native, fully oxidized b 5 R family members in which the nicotinamide ring of NAD(P) + resides in the active site. Finally, the structures, together with sequence alignments, show that the b 5 R domain is more closely related to single-domain Cyb5R proteins from plants, fungi and some protists than to Cyb5R3 from animals.
Organizational Affiliation:
Department of Chemistry, The University of Kansas, 1567 Irving Hill Road, Lawrence, KS 66045, USA.