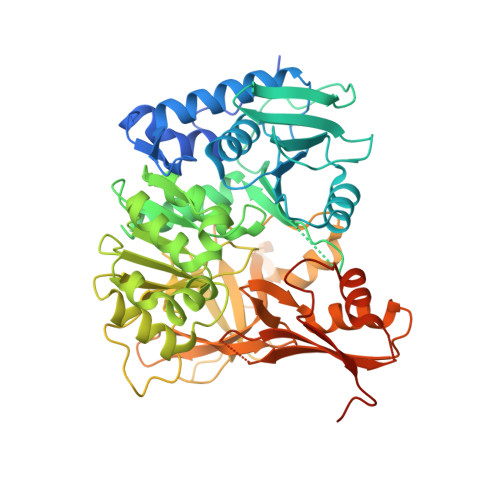
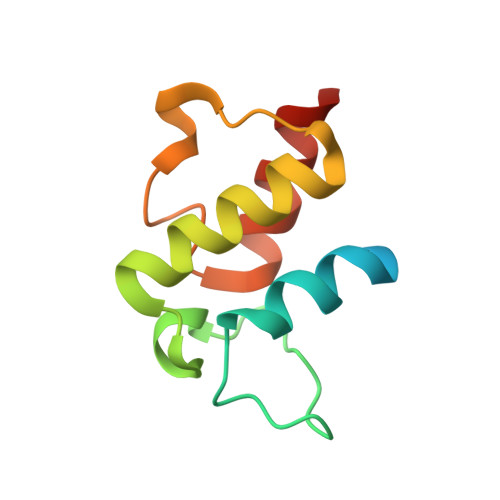
Structural Characterization of Complex of Adenylation Domain and Carrier Protein by Using Pantetheine Cross-Linking Probe.
Miyanaga, A., Kurihara, S., Chisuga, T., Kudo, F., Eguchi, T.(2020) ACS Chem Biol 15: 1808-1812
- PubMed: 32608966
- DOI: https://doi.org/10.1021/acschembio.0c00403
- Primary Citation of Related Structures:
6M01 - PubMed Abstract:
Adenylation domains (A-domains) are responsible for selective incorporation of carboxylic acid substrates in the biosynthesis of various natural products. Each A-domain must recognize a cognate carrier protein (CP) for functional substrate transfer. The transient interactions between an A-domain and CP have been investigated by using acyl vinylsulfonamide adenosine inhibitors as probes to determine the structures of several A-domain-CP complexes. However, this strategy requires a specific vinylsulfonamide inhibitor that contains an acyl group corresponding to the substrate specificity of a target A-domain in every case. Here, we report an alternative strategy for structural characterization of A-domain-CP complexes. We used a bromoacetamide pantetheine cross-linking probe in combination with a Cys mutation to trap the standalone A-domain-CP complex involved in macrolactam polyketide biosynthesis through a covalent linkage, allowing the determination of the complex structure. This strategy facilitates the structural determination of A-domain-CP complexes.
Organizational Affiliation:
Department of Chemistry, Tokyo Institute of Technology, 2-12-1 Meguro-ku, O-okayama, Tokyo 152-8551, Japan.