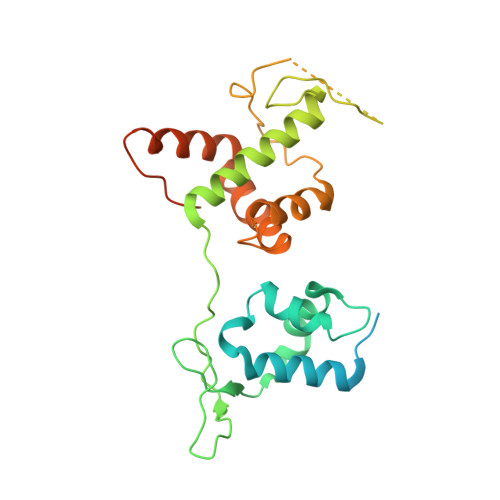
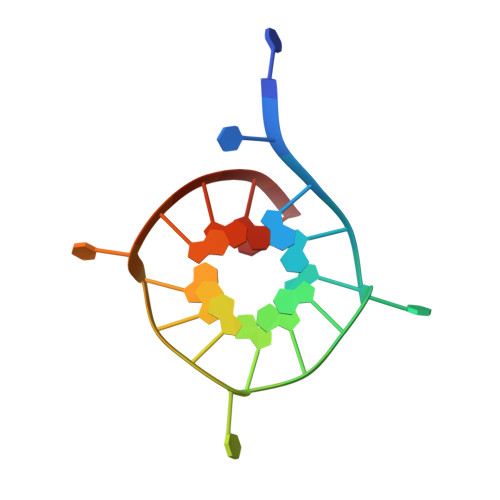
Structural basis of G-quadruplex DNA recognition by the yeast telomeric protein Rap1.
Traczyk, A., Liew, C.W., Gill, D.J., Rhodes, D.(2020) Nucleic Acids Res 48: 4562-4571
- PubMed: 32187364
- DOI: https://doi.org/10.1093/nar/gkaa171
- Primary Citation of Related Structures:
6LDM - PubMed Abstract:
G-quadruplexes are four-stranded nucleic acid structures involved in multiple cellular pathways including DNA replication and telomere maintenance. Such structures are formed by G-rich DNA sequences typified by telomeric DNA repeats. Whilst there is evidence for proteins that bind and regulate G-quadruplex formation, the molecular basis for this remains poorly understood. The budding yeast telomeric protein Rap1, originally identified as a transcriptional regulator functioning by recognizing double-stranded DNA binding sites, was one of the first proteins to be discovered to also bind and promote G-quadruplex formation in vitro. Here, we present the 2.4 Å resolution crystal structure of the Rap1 DNA-binding domain in complex with a G-quadruplex. Our structure not only provides a detailed insight into the structural basis for G-quadruplex recognition by a protein, but also gives a mechanistic understanding of how the same DNA-binding domain adapts to specifically recognize different DNA structures. The key observation is the DNA-recognition helix functions in a bimodal manner: In double-stranded DNA recognition one helix face makes electrostatic interactions with the major groove of DNA, whereas in G-quadruplex recognition a different helix face is used to make primarily hydrophobic interactions with the planar face of a G-tetrad.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University Singapore, 60 Nanyang Drive, Singapore 637551, Singapore.