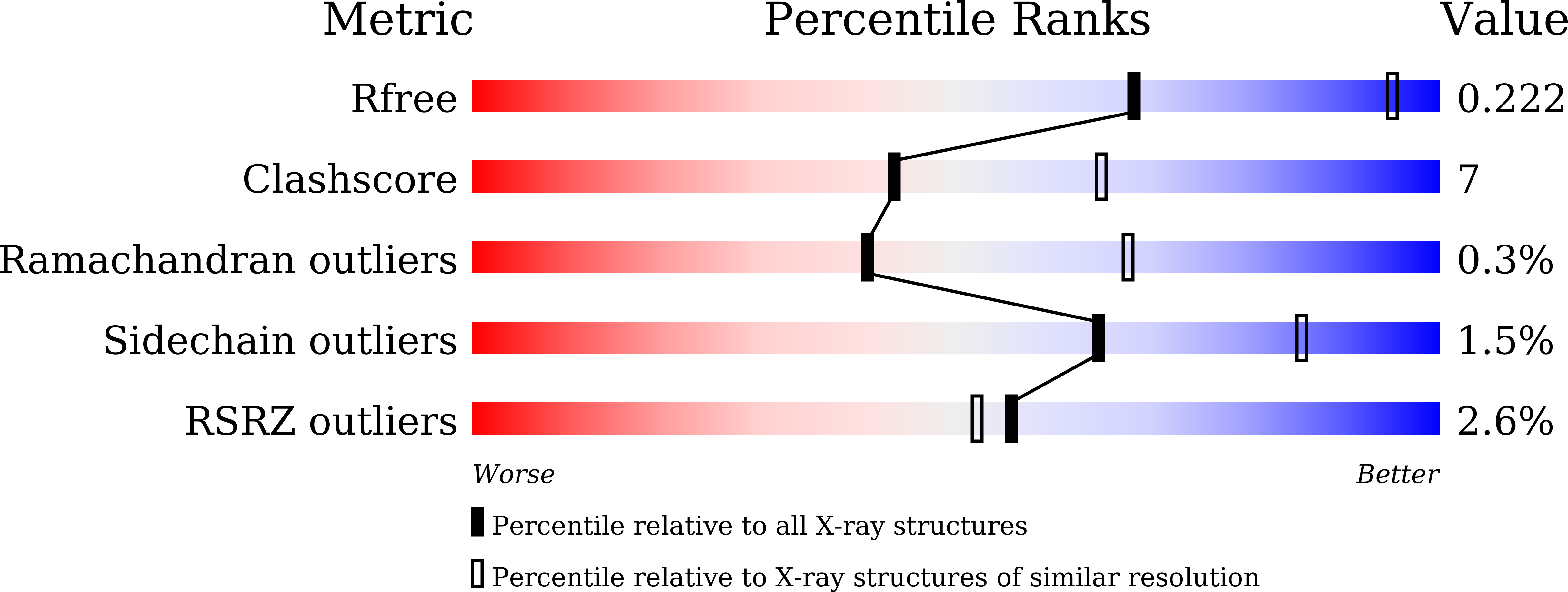
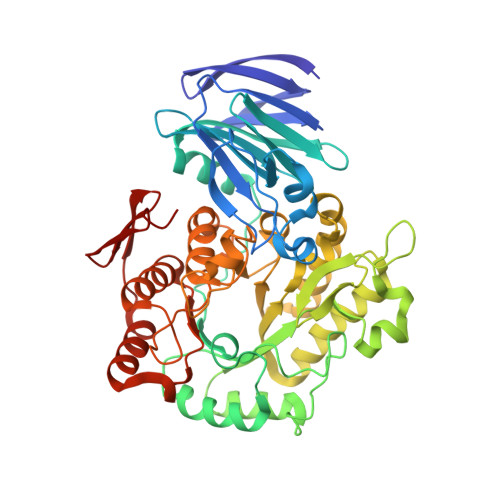
Crystal Structure of alpha-Galactosidase fromThermus thermophilus: Insight into Hexamer Assembly and Substrate Specificity.
Chen, S.C., Wu, S.P., Chang, Y.Y., Hwang, T.S., Lee, T.H., Hsu, C.H.(2020) J Agric Food Chem 68: 6161-6169
- PubMed: 32390413
- DOI: https://doi.org/10.1021/acs.jafc.0c00871
- Primary Citation of Related Structures:
6LCJ, 6LCK, 6LCL - PubMed Abstract:
α-Galactosidase catalyzes the hydrolysis of a terminal α-galactose residue in galacto-oligosaccharides and has potential in various industrial applications and food processing. We determined the crystal structures of α-galactosidase from the thermophilic microorganism Thermus thermophilus (TtGalA) and its complexes with pNPGal and stachyose. The monomer folds into an N-terminal domain, a catalytic (β/α) 8 barrel domain, and a C-terminal domain. The domain organization is similar to that of the other family of 36 α-galactosidases, but TtGalA presents a cagelike hexamer. Structural analysis shows that oligomerization may be a key factor for the thermal adaption of TtGalA. The structure of TtGalA complexed with stachyose reveals only the existence of one -1 subsite and one +1 subsite in the active site. Structural comparison of the stachyose-bound complexes of TtGalA and GsAgaA, a tetrameric enzyme with four subsites, suggests evolutionary divergence of substrate specificity within the GH36 family of α-galactosidases. To the best of our knowledge, the crystal structure of TtGalA is the first report of a quaternary structure as a hexameric assembly in the α-galactosidase family.
Organizational Affiliation:
Department of Agricultural Chemistry, National Taiwan University, Taipei 10617, Taiwan.