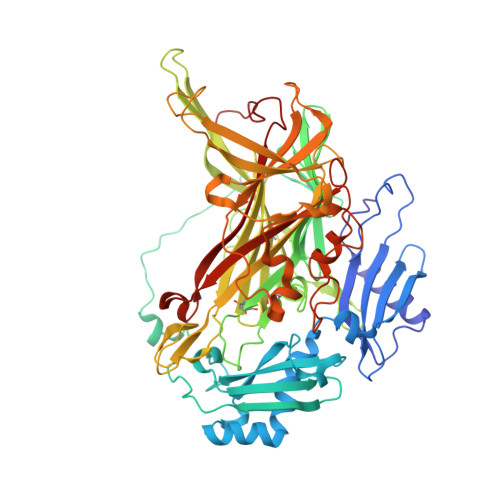
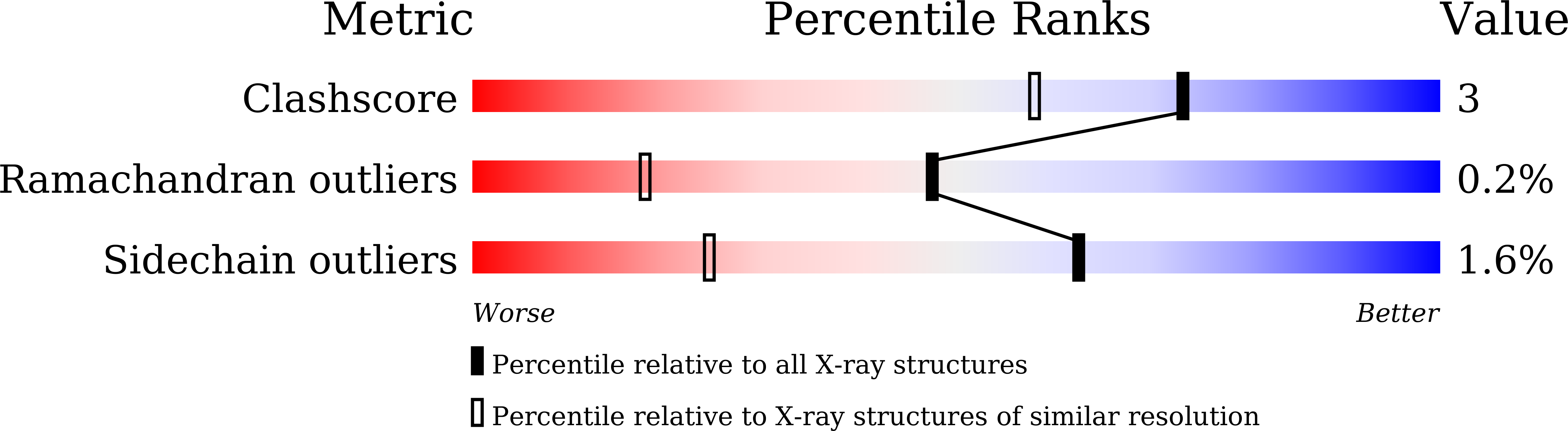Neutron crystallography of copper amine oxidase reveals keto/enolate interconversion of the quinone cofactor and unusual proton sharing.
Murakawa, T., Kurihara, K., Shoji, M., Shibazaki, C., Sunami, T., Tamada, T., Yano, N., Yamada, T., Kusaka, K., Suzuki, M., Shigeta, Y., Kuroki, R., Hayashi, H., Yano, T., Tanizawa, K., Adachi, M., Okajima, T.(2020) Proc Natl Acad Sci U S A 117: 10818-10824
- PubMed: 32371483
- DOI: https://doi.org/10.1073/pnas.1922538117
- Primary Citation of Related Structures:
6L9C - PubMed Abstract:
Recent advances in neutron crystallographic studies have provided structural bases for quantum behaviors of protons observed in enzymatic reactions. Thus, we resolved the neutron crystal structure of a bacterial copper (Cu) amine oxidase (CAO), which contains a prosthetic Cu ion and a protein-derived redox cofactor, topa quinone (TPQ). We solved hitherto unknown structures of the active site, including a keto/enolate equilibrium of the cofactor with a nonplanar quinone ring, unusual proton sharing between the cofactor and the catalytic base, and metal-induced deprotonation of a histidine residue that coordinates to the Cu. Our findings show a refined active-site structure that gives detailed information on the protonation state of dissociable groups, such as the quinone cofactor, which are critical for catalytic reactions.
Organizational Affiliation:
Department of Biochemistry, Osaka Medical College, 569-8686 Takatsuki, Osaka, Japan; murakawa@osaka-med.ac.jp tokajima@sanken.osaka-u.ac.jp.