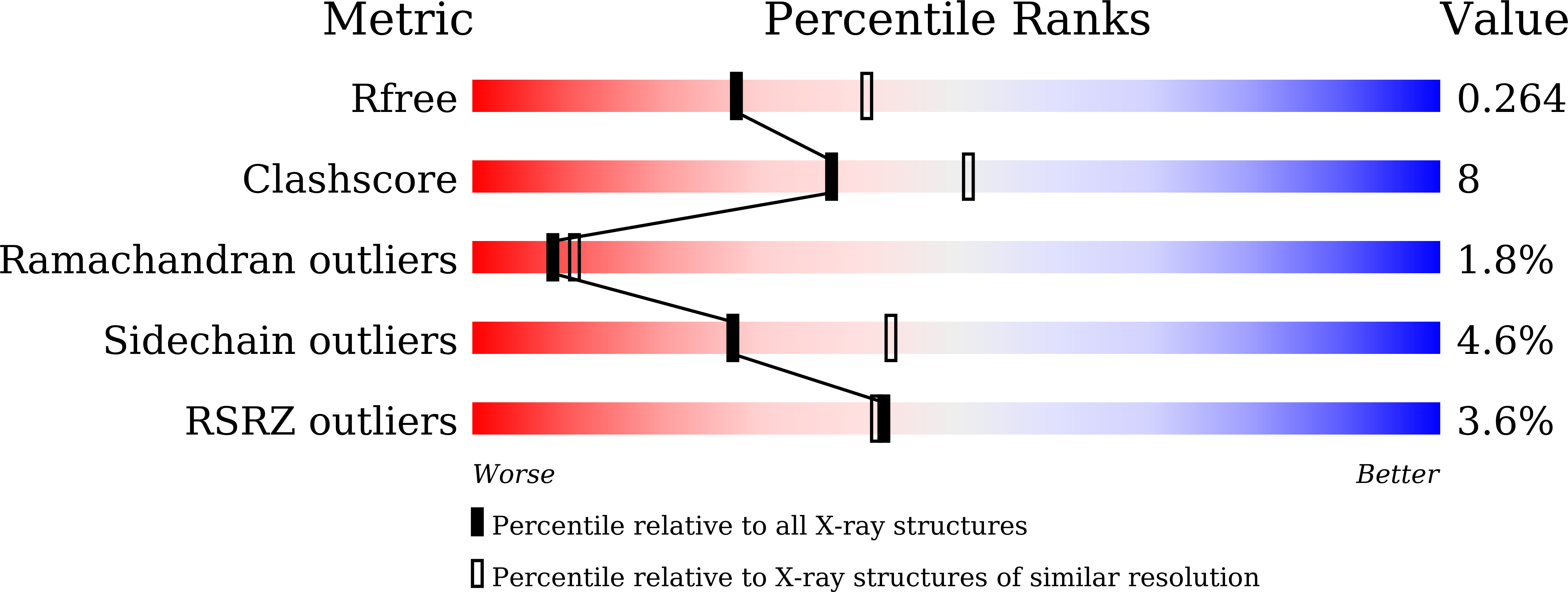
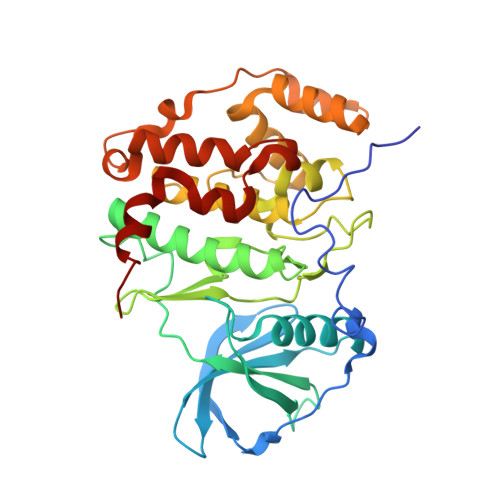
Structural insights for producing CK2 alpha 1-specific inhibitors.
Tsuyuguchi, M., Nakaniwa, T., Hirasawa, A., Nakanishi, I., Kinoshita, T.(2020) Bioorg Med Chem Lett 30: 126837-126837
- PubMed: 31859160
- DOI: https://doi.org/10.1016/j.bmcl.2019.126837
- Primary Citation of Related Structures:
6L1Z, 6L20, 6L21, 6L22, 6L23, 6L24 - PubMed Abstract:
Casein kinase 2 catalytic subunit (CK2α) is classified into two subtypes CK2α1 and CK2α2. CK2α1 is a drug discovery target, whereas CK2α2 is an off-target of CK2α1 inhibitors. High amino acid sequence homology between these subtypes hampers efforts to produce ATP competitive inhibitors that are highly selective to CK2α1. Hematein was identified previously as a non-ATP-competitive inhibitor for CK2α1, whereas this compound acts as an ATP competitive CK2α2 inhibitor. Crystal structures of CK2α1 and CK2α2 in complex with hematein revealed distinct binding features that provide structural insights for producing CK2α1-selective inhibitors.
Organizational Affiliation:
Graduate School of Science, Osaka Prefecture University, 1-1 Gakuen-cho, Naka-ku, Sakai, Osaka 599-8531, Japan. Electronic address: tsuyuguchi13@b.s.osakafu-u.ac.jp.