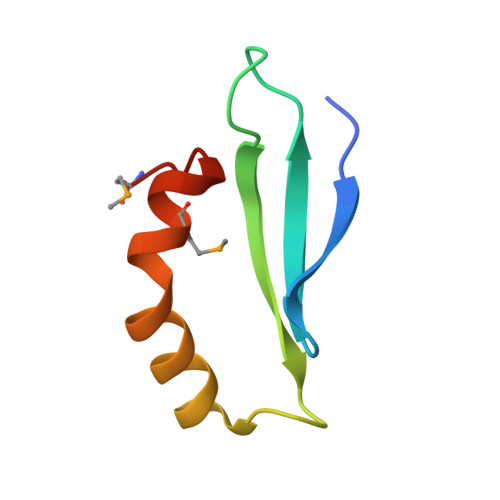
Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control.
Furutani, M., Hirano, Y., Nishimura, T., Nakamura, M., Taniguchi, M., Suzuki, K., Oshida, R., Kondo, C., Sun, S., Kato, K., Fukao, Y., Hakoshima, T., Morita, M.T.(2020) Nat Commun 11: 76-76
- PubMed: 31900388
- DOI: https://doi.org/10.1038/s41467-019-13729-7
- Primary Citation of Related Structures:
6L0V, 6L0W - PubMed Abstract:
In many plant species, roots maintain specific growth angles relative to the direction of gravity, known as gravitropic set point angles (GSAs). These contribute to the efficient acquisition of water and nutrients. AtLAZY1/LAZY1-LIKE (LZY) genes are involved in GSA control by regulating auxin flow toward the direction of gravity in Arabidopsis. Here, we demonstrate that RCC1-like domain (RLD) proteins, identified as LZY interactors, are essential regulators of polar auxin transport. We show that interaction of the CCL domain of LZY with the BRX domain of RLD is important for the recruitment of RLD from the cytoplasm to the plasma membrane by LZY. A structural analysis reveals the mode of the interaction as an intermolecular β-sheet in addition to the structure of the BRX domain. Our results offer a molecular framework in which gravity signal first emerges as polarized LZY3 localization in gravity-sensing cells, followed by polar RLD1 localization and PIN3 relocalization to modulate auxin flow.
Organizational Affiliation:
College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, Fujian, 350002, China.