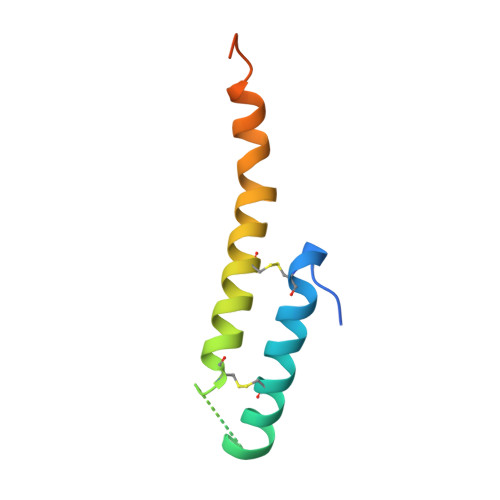
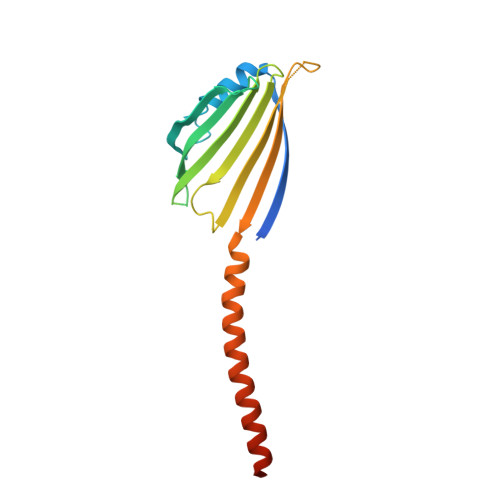
Molecular mechanism of mitochondrial phosphatidate transfer by Ups1.
Lu, J., Chan, C., Yu, L., Fan, J., Sun, F., Zhai, Y.(2020) Commun Biol 3: 468-468
- PubMed: 32843686
- DOI: https://doi.org/10.1038/s42003-020-01121-x
- Primary Citation of Related Structures:
6KYL - PubMed Abstract:
Cardiolipin, an essential mitochondrial physiological regulator, is synthesized from phosphatidic acid (PA) in the inner mitochondrial membrane (IMM). PA is synthesized in the endoplasmic reticulum and transferred to the IMM via the outer mitochondrial membrane (OMM) under mediation by the Ups1/Mdm35 protein family. Despite the availability of numerous crystal structures, the detailed mechanism underlying PA transfer between mitochondrial membranes remains unclear. Here, a model of Ups1/Mdm35-membrane interaction is established using combined crystallographic data, all-atom molecular dynamics simulations, extensive structural comparisons, and biophysical assays. The α2-loop, L2-loop, and α3 helix of Ups1 mediate membrane interactions. Moreover, non-complexed Ups1 on membranes is found to be a key transition state for PA transfer. The membrane-bound non-complexed Ups1/ membrane-bound Ups1 ratio, which can be regulated by environmental pH, is inversely correlated with the PA transfer activity of Ups1/Mdm35. These results demonstrate a new model of the fine conformational changes of Ups1/Mdm35 during PA transfer.
Organizational Affiliation:
National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, 100101, Beijing, China.