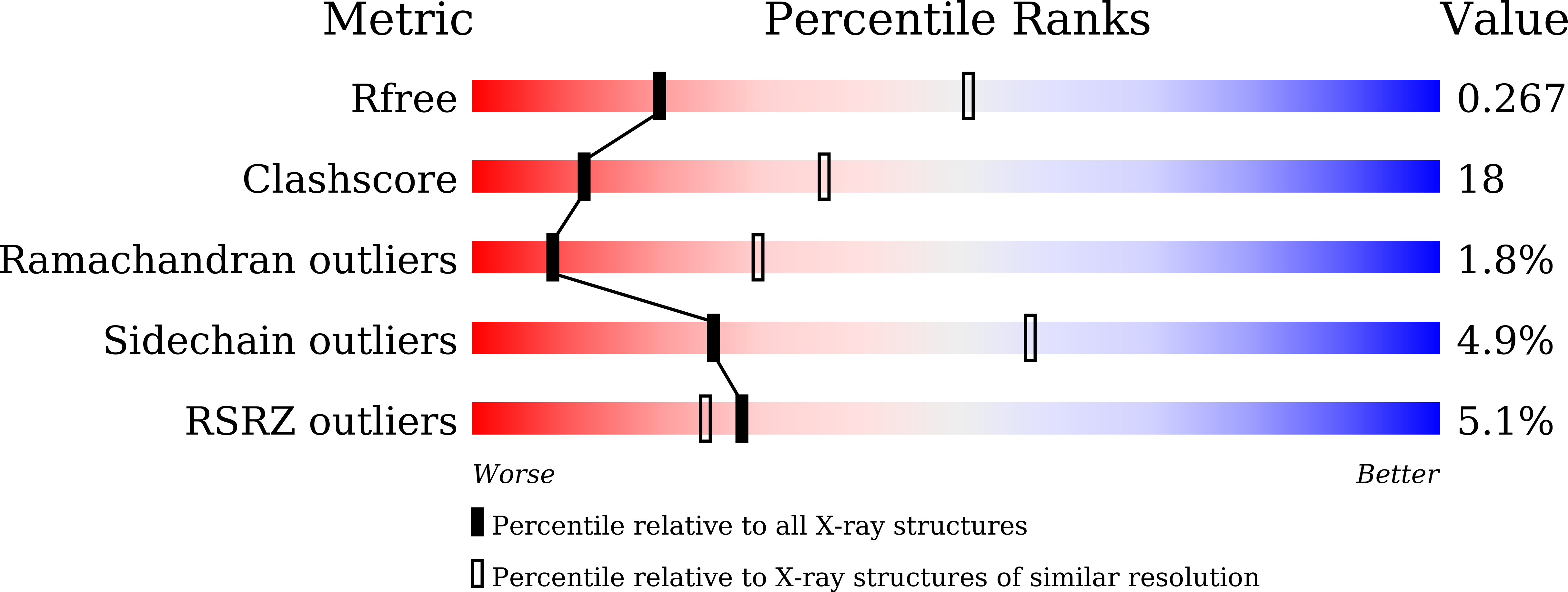
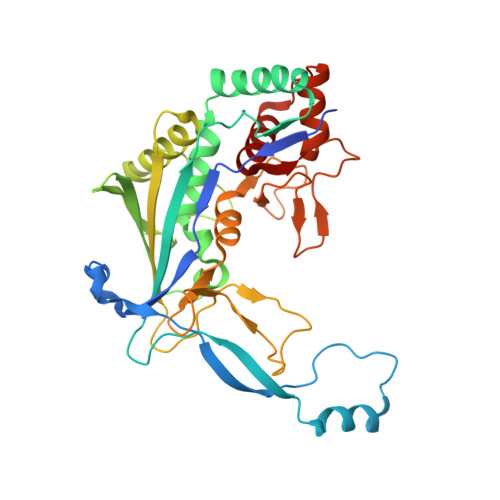
A CRISPR RNA Is Closely Related With the Size of the Cascade Nucleoprotein Complex.
Gu, D.H., Ha, S.C., Kim, J.S.(2019) Front Microbiol 10: 2458-2458
- PubMed: 31736904
- DOI: https://doi.org/10.3389/fmicb.2019.02458
- Primary Citation of Related Structures:
6KQR - PubMed Abstract:
The currently known prokaryotic adaptive immune system against mobile genetic elements is based on clustered regularly interspaced short palindromic repeats (CRISPR). CRISPR-associated (Cas) proteins and the transcribed short CRISPR RNA (crRNA) molecule form a heterologous ribonucleoprotein complex that neutralizes invading foreign nucleic acids, wherein the crRNA molecule base-pairs with the exogenous genetic elements. In the ribonucleoprotein complexes of the type I CRISPR system, a helical backbone of six identical subunits is commonly found. However, it is not clear how this ribonucleoprotein complex is assembled and what is the determinant factor for its size. We elucidated the crystal structure of the Csy3 subunit of the type I-F ribonucleoprotein complex from Zymomonas mobilis (ZmCsy3), in which seven ZmCsy3 protomers in the asymmetric unit form a molecular helix that is part of a filamentous structure in the entire crystal system. This ZmCsy3 helical structure is remarkably similar to the crRNA-bound hexameric Csy3 backbone from Pseudomonas aeruginosa , with conserved interactions between neighboring subunits. The monomeric ZmCsy3 in solution is transformed into different oligomeric states depending on the added crRNAs. These results suggest that a crRNA and Csy3 subunit play a determinant role in the stepwise formation of the functional Cascade ribonucleoprotein complex and the recruitment of other subunits, and crRNA functions as a molecular ruler for determining the size of the Cascade silencing complex.
Organizational Affiliation:
Department of Chemistry, Chonnam National University, Gwangju, South Korea.