NAG-thiazoline is a potent inhibitor of the Vibrio campbellii GH20 beta-N-Acetylglucosaminidase.
Meekrathok, P., Stubbs, K.A., Aunkham, A., Kaewmaneewat, A., Kardkuntod, A., Bulmer, D.M., van den Berg, B., Suginta, W.(2020) FEBS J 287: 4982-4995
- PubMed: 32145141
- DOI: https://doi.org/10.1111/febs.15283
- Primary Citation of Related Structures:
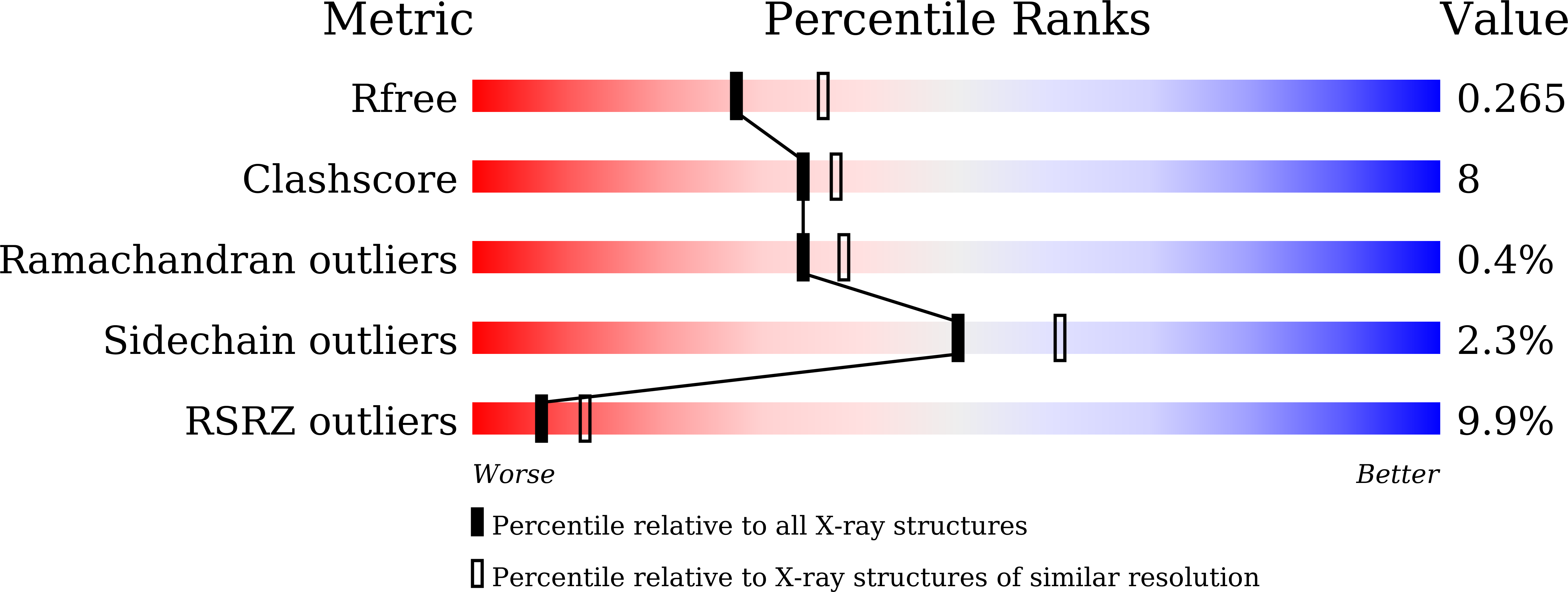
6K35 - PubMed Abstract:
Vibrio spp. play a vital role in the recycling of chitin in oceans, but several Vibrio strains are highly infectious to aquatic animals and humans. These bacteria require chitin for growth; thus, potent inhibitors of chitin-degrading enzymes could serve as candidate drugs against Vibrio infections. This study examined NAG-thiazoline (NGT)-mediated inhibition of a recombinantly expressed GH20 β-N-acetylglucosaminidase, namely VhGlcNAcase from Vibrio campbellii (formerly V. harveyi) ATCC BAA-1116. NGT strongly inhibited VhGlcNAcase with an IC 50 of 11.9 ± 1.0 μm and K i 62 ± 3 µm, respectively. NGT was also found to completely inhibit the growth of V. campbellii strain 650 with an minimal inhibitory concentration value of 0.5 µm. ITC data analysis showed direct binding of NGT to VhGlcNAcase with a K d of 32 ± 1.2 μm. The observed ΔG° binding of -7.56 kcal·mol -1 is the result of a large negative enthalpy change and a small positive entropic compensation, suggesting that NGT binding is enthalpy-driven. The structural complex shows that NGT fully occupies the substrate-binding pocket of VhGlcNAcase and makes an exclusive hydrogen bond network, as well as hydrophobic interactions with the conserved residues around the -1 subsite. Our results strongly suggest that NGT could serve as an excellent scaffold for further development of antimicrobial agents against Vibrio infections. DATABASE: Structural data are available in PDB database under the accession number 6K35.
Organizational Affiliation:
School of Chemistry, Suranaree University of Technology, Nakhon Ratchasima, Thailand.