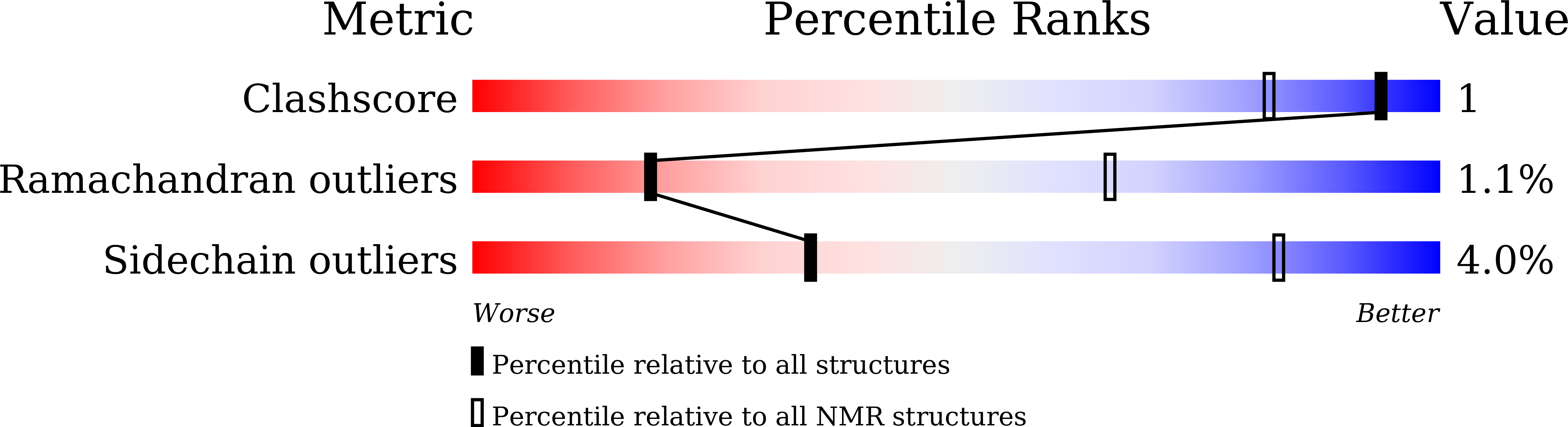
Chemical shift assignments of a camelid nanobody against aflatoxin B1.
Nie, Y., Li, S., Zhu, J., Hu, R., Liu, M., He, T., Yang, Y.(2019) Biomol NMR Assign 13: 75-78
- PubMed: 30328057
- DOI: https://doi.org/10.1007/s12104-018-9855-y
- Primary Citation of Related Structures:
6IYN - PubMed Abstract:
Nanobodies (Nbs) are the variable domain of the heavy-chain antibodies produced from Camelidae, which possess comparable binding affinities and specificity to conventional antibodies. Nbs have become valuable and versatile tools for numerous biotechnology applications due to their small size (12-15 kDa), high solubility, exceptional stability, and facile genetic manipulation. The interactions between Nbs and protein antigens have been well-studied, but less work has been done to characterize their ability to bind small molecule haptens. Here we present the backbone and side-chain assignments of the 1 H, 13 C and 15 N resonances of Nb26 (123 amino acids), a nanobody that recognizes the hapten aflatoxin B 1 (AFB 1 ). These assignments are preliminary work towards the determination of the structure of free Nb26 using NMR spectroscopy, which will provide useful information about the complex structure of "Nb26-AFB 1 " and the recognition mechanism about how Nb26 binds to AFB 1 .
Organizational Affiliation:
State Key Laboratory of Magnetic Resonance and Atomic Molecular Physics, Wuhan Center for Magnetic Resonance, Wuhan Institute of Physics and Mathematics, Chinese Academy of Sciences, Wuhan, 430071, China.