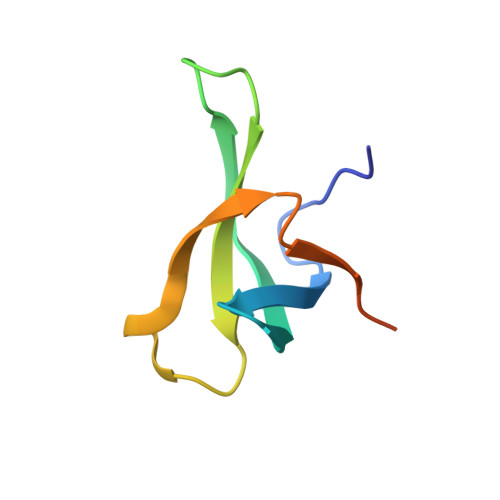
Crystal structure of TbEsa1 presumed Tudor domain from Trypanosoma brucei.
Gao, J., Ye, K., Diwu, Y., Xu, C., Zhang, X., Liao, S., Tu, X.(2020) J Struct Biol 209: 107406-107406
- PubMed: 31747559
- DOI: https://doi.org/10.1016/j.jsb.2019.107406
- Primary Citation of Related Structures:
6IF4 - PubMed Abstract:
The essential SAS2-related acetyltransferase 1 (Esa1), as a acetyltransferase of MYST family, is indispensable for the cell cycle and transcriptional regulation. The Tudor domain consists of 60 amino acids and belongs to the Royal family, which serves as a module interacting with methylated histone and/or DNA. Although Tudor domain has been widely studied in higher eukaryotes, its structure and function remain unclear in Trypanosoma brucei (T. brucei), a protozoan unicellular parasite causing sleeping sickness in human and nagana in cattle in sub-Saharan Africa. Here, we determined a high-resolution structure of TbEsa1 presumed Tudor domain from T. brucei by X-ray crystallography. TbEsa1 Tudor domain adopts a conserved Tudor-like fold, which is comprised of a five-stranded β-barrel surrounded by two short α-helices. Furthermore, we revealed a non-specific DNA binding pattern of TbEsa1 Tudor domain. However, TbEsa1 Tudor domain showed no methyl-histone binding ability, due to the absence of key aromatic residues forming a conserved aromatic cage.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at Microscale, and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui, PR China.