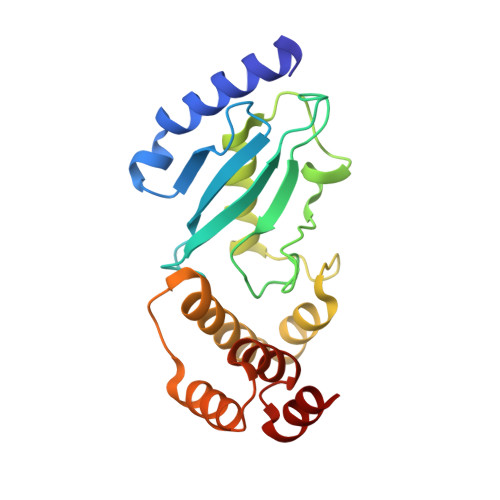
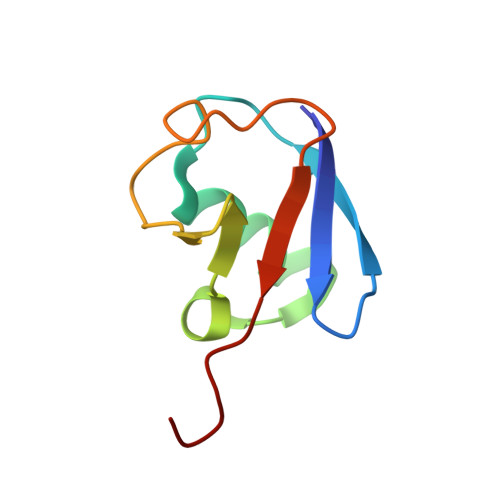
Crystal structure of the Ube2K/E2-25K and K48-linked di-ubiquitin complex provides structural insight into the mechanism of K48-specific ubiquitin chain synthesis.
Lee, J.G., Youn, H.S., Kang, J.Y., Park, S.Y., Kidera, A., Yoo, Y.J., Eom, S.H.(2018) Biochem Biophys Res Commun 506: 102-107
- PubMed: 30336976
- DOI: https://doi.org/10.1016/j.bbrc.2018.10.067
- Primary Citation of Related Structures:
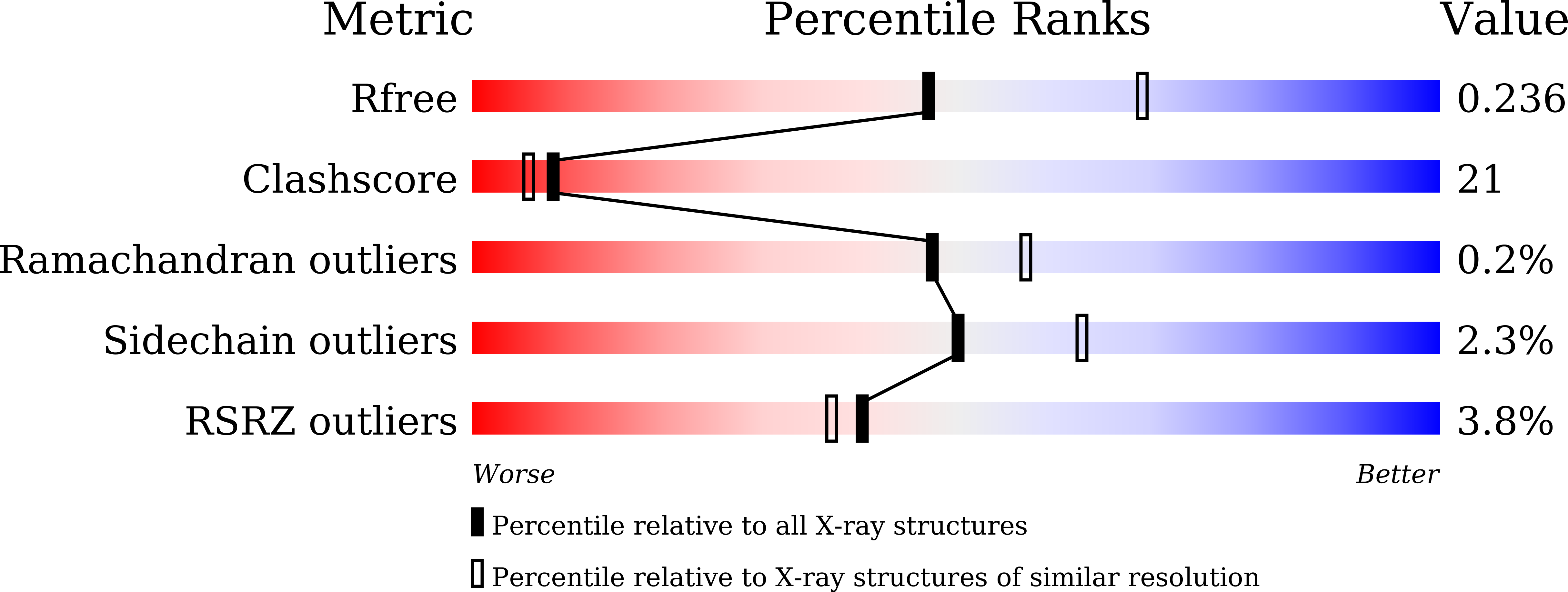
6IF1 - PubMed Abstract:
Ubiquitin-conjugating enzymes (E2) form thioester bonds with ubiquitin (Ub), which are subsequently transferred to target proteins for cellular progress. Ube2K/E2-25K (a class II E2 enzyme) contains a C-terminal ubiquitin-associated (UBA) domain that has been suggested to control ubiquitin recognition, dimerization, or poly-ubiquitin chain formation. Ube2K is a special E2 because it synthesizes K48-linked poly-ubiquitin chains without E3 ubiquitin ligase. We found that a novel interaction between the acceptor di-Ub (Ub 2 ) and the auxiliary Ube2K promotes the discharging reaction and production of tri-Ub (Ub 3 ), probably by guiding and positioning the K48 (in the distal Ub) of the acceptor Ub 2 in the active site. We also determined the crystal structure of Ube2K-Ub 2 at 2.47 Å resolution. Based on our structural and biochemical data, we proposed a structural model of Ub 3 synthesis by Ube2K without E3.
Organizational Affiliation:
School of Life Sciences, Gwangju Institute of Science and Technology, Gwangju, 61005, Republic of Korea; Steitz Center for Structural Biology, Gwangju Institute of Science and Technology, Gwangju, 61005, Republic of Korea.