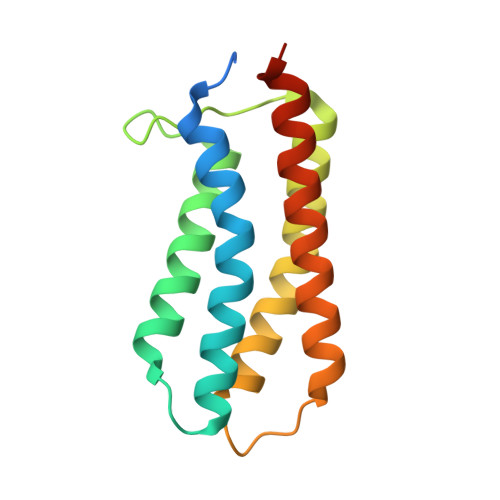
The Molecular Mechanism of Nitrate Chemotaxis via Direct Ligand Binding to the PilJ Domain of McpN.
Martin-Mora, D., Ortega, A., Matilla, M.A., Martinez-Rodriguez, S., Gavira, J.A., Krell, T.(2019) mBio 10
- PubMed: 30782655
- DOI: https://doi.org/10.1128/mBio.02334-18
- Primary Citation of Related Structures:
6GCV - PubMed Abstract:
Chemotaxis and energy taxis permit directed bacterial movements in gradients of environmental cues. Nitrate is a final electron acceptor for anaerobic respiration and can also serve as a nitrogen source for aerobic growth. Previous studies indicated that bacterial nitrate taxis is mediated by energy taxis mechanisms, which are based on the cytosolic detection of consequences of nitrate metabolism. Here we show that Pseudomonas aeruginosa PAO1 mediates nitrate chemotaxis on the basis of specific nitrate sensing by the periplasmic PilJ domain of the PA2788/McpN chemoreceptor. The presence of nitrate reduced mcpN transcript levels, and McpN-mediated taxis occurred only under nitrate starvation conditions. In contrast to the NarX and NarQ sensor kinases, McpN bound nitrate specifically and showed no affinity for other ligands such as nitrite. We report the three-dimensional structure of the McpN ligand binding domain (LBD) at 1.3-Å resolution in complex with nitrate. Although structurally similar to 4-helix bundle domains, the ligand binding mode differs since a single nitrate molecule is bound to a site on the dimer symmetry axis. As for 4-helix bundle domains, ligand binding stabilized the McpN-LBD dimer. McpN homologues showed a wide phylogenetic distribution, indicating that nitrate chemotaxis is a widespread phenotype. These homologues were particularly abundant in bacteria that couple sulfide/sulfur oxidation with nitrate reduction. This work expands the range of known chemotaxis effectors and forms the basis for the exploration of nitrate chemotaxis in other bacteria and for the study of its physiological role. IMPORTANCE Nitrate is of central importance in bacterial physiology. Previous studies indicated that movements toward nitrate are due to energy taxis, which is based on the cytosolic sensing of consequences of nitrate metabolism. Here we present the first report on nitrate chemotaxis. This process is initiated by specific nitrate binding to the periplasmic ligand binding domain (LBD) of McpN. Nitrate chemotaxis is highly regulated and occurred only under nitrate starvation conditions, which is helpful information to explore nitrate chemotaxis in other bacteria. We present the three-dimensional structure of the McpN-LBD in complex with nitrate, which is the first structure of a chemoreceptor PilJ-type domain. This structure reveals striking similarities to that of the abundant 4-helix bundle domain but employs a different sensing mechanism. Since McpN homologues show a wide phylogenetic distribution, nitrate chemotaxis is likely a widespread phenomenon with importance for the life cycle of ecologically diverse bacteria.
Organizational Affiliation:
Estación Experimental del Zaidín, Department of Environmental Protection, Consejo Superior de Investigaciones Científicas, Granada, Spain.