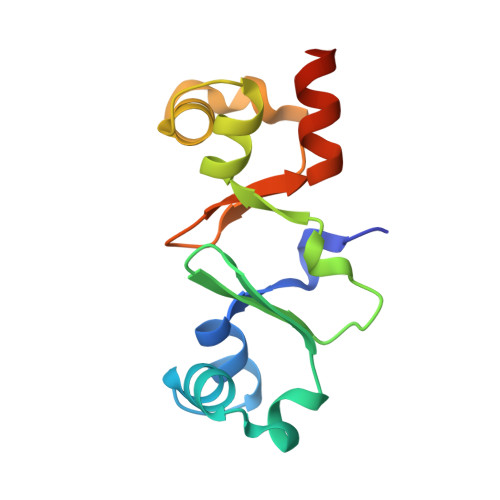
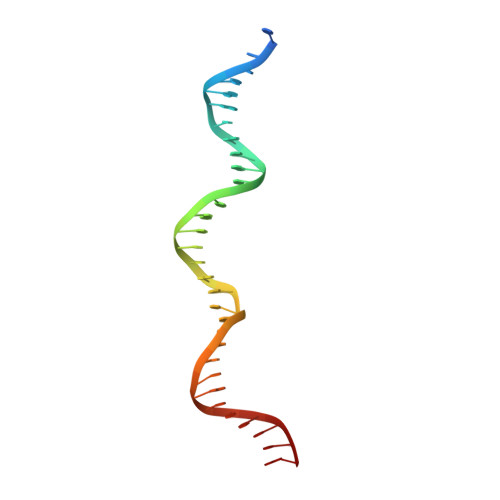
TraN: A novel repressor of an Enterococcus conjugative type IV secretion system.
Kohler, V., Goessweiner-Mohr, N., Aufschnaiter, A., Fercher, C., Probst, I., Pavkov-Keller, T., Hunger, K., Wolinski, H., Buttner, S., Grohmann, E., Keller, W.(2018) Nucleic Acids Res 46: 9201-9219
- PubMed: 30060171
- DOI: https://doi.org/10.1093/nar/gky671
- Primary Citation of Related Structures:
6G1T - PubMed Abstract:
The dissemination of multi-resistant bacteria represents an enormous burden on modern healthcare. Plasmid-borne conjugative transfer is the most prevalent mechanism, requiring a type IV secretion system that enables bacteria to spread beneficial traits, such as resistance to last-line antibiotics, among different genera. Inc18 plasmids, like the Gram-positive broad host-range plasmid pIP501, are substantially involved in propagation of vancomycin resistance from Enterococci to methicillin-resistant strains of Staphylococcus aureus. Here, we identified the small cytosolic protein TraN as a repressor of the pIP501-encoded conjugative transfer system, since deletion of traN resulted in upregulation of transfer factors, leading to highly enhanced conjugative transfer. Furthermore, we report the complex structure of TraN with DNA and define the exact sequence of its binding motif. Targeting this protein-DNA interaction might represent a novel therapeutic approach against the spreading of antibiotic resistances.
Organizational Affiliation:
Institute of Molecular Biosciences, University of Graz, Graz 8010, Austria.