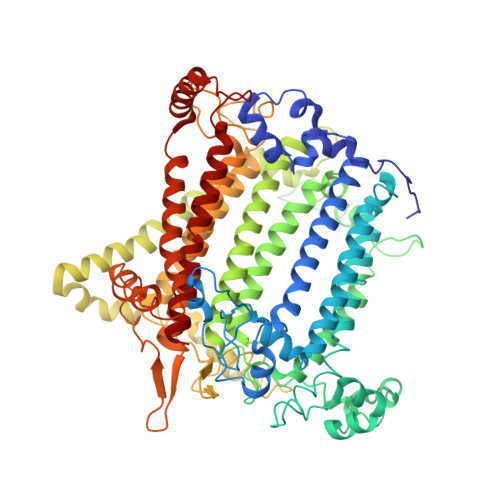
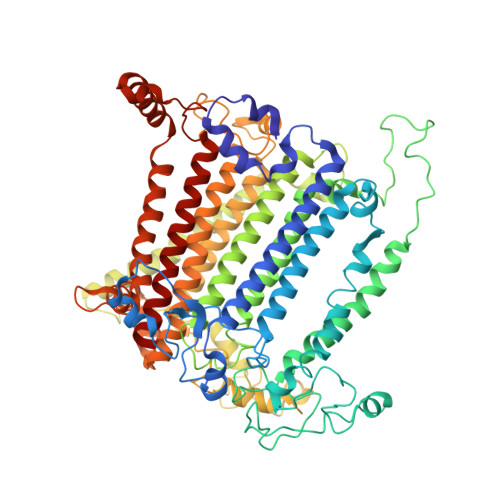
Structure and function of photosystem I in Cyanidioschyzon merolae.
Antoshvili, M., Caspy, I., Hippler, M., Nelson, N.(2019) Photosynth Res 139: 499-508
- PubMed: 29582227
- DOI: https://doi.org/10.1007/s11120-018-0501-4
- Primary Citation of Related Structures:
6FOS - PubMed Abstract:
The evolution of photosynthesis from primitive photosynthetic bacteria to higher plants has been driven by the need to adapt to a wide range of environmental conditions. The red alga Cyanidioschyzon merolae is a primitive organism, which is capable of performing photosynthesis in extreme acidic and hot environments. The study of its photosynthetic machinery may provide new insight on the evolutionary path of photosynthesis and on light harvesting and its regulation in eukaryotes. With that aim, the structural and functional properties of the PSI complex were investigated by biochemical characterization, mass spectrometry, and X-ray crystallography. PSI was purified from cells grown at 25 and 42 °C, crystallized and its crystal structure was solved at 4 Å resolution. The structure of C. merolae reveals a core complex with a crescent-shaped structure, formed by antenna proteins. In addition, the structural model shows the position of PsaO and PsaM. PsaG and PsaH are present in plant complex and are missing from the C. merolae model as expected. This paper sheds new light onto the evolution of photosynthesis, which gives a strong indication for the chimerical properties of red algae PSI. The subunit composition of the PSI core from C. merolae and its associated light-harvesting antennae suggests that it is an evolutionary and functional intermediate between cyanobacteria and plants.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The George S. Wise Faculty of Life Sciences, Tel Aviv University, 69978, Tel Aviv, Israel.