Identification of Noncatalytic Lysine Residues from Allosteric Circuits via Covalent Probes.
Bongard, J., Lorenz, M., Vetter, I.R., Stege, P., Porfetye, A.T., Schmitz, A.L., Kaschani, F., Wolf, A., Koch, U., Nussbaumer, P., Klebl, B., Kaiser, M., Ehrmann, M.(2018) ACS Chem Biol 13: 1307-1312
- PubMed: 29658704
- DOI: https://doi.org/10.1021/acschembio.8b00101
- Primary Citation of Related Structures:
6EW9 - PubMed Abstract:
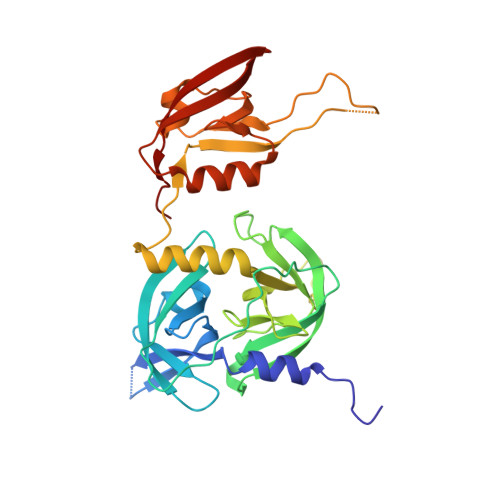
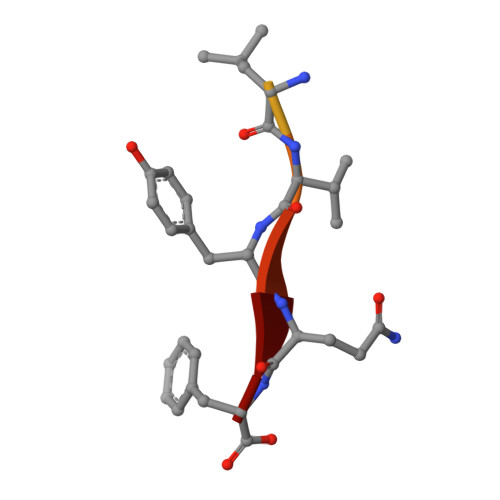
Covalent modifications of nonactive site lysine residues by small molecule probes has recently evolved into an important strategy for interrogating biological systems. Here, we report the discovery of a class of bioreactive compounds that covalently modify lysine residues in DegS, the rate limiting protease of the essential bacterial outer membrane stress response pathway. These modifications lead to an allosteric activation and allow the identification of novel residues involved in the allosteric activation circuit. These findings were validated by structural analyses via X-ray crystallography and cell-based reporter systems. We anticipate that our findings are not only relevant for a deeper understanding of the structural basis of allosteric activation in DegS and other HtrA serine proteases but also pinpoint an alternative use of covalent small molecules for probing essential biochemical mechanisms.
Organizational Affiliation:
Microbiology, Faculty of Biology, Zentrum für Medizinische Biotechnologie (ZMB) , Universität Duisburg-Essen , Universitätsstr. 2 , 45117 Essen , Germany.