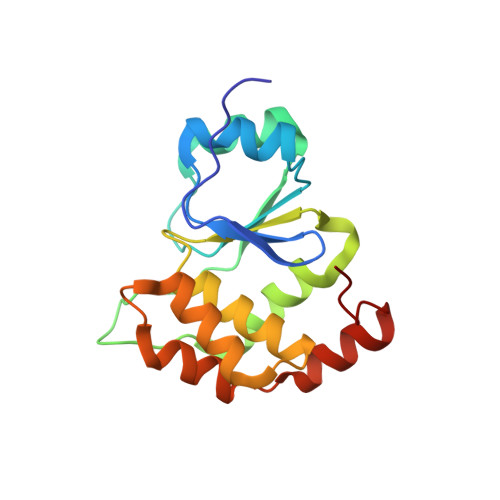
Molecular Architecture of the Inositol Phosphatase Siw14.
Florio, T.J., Lokareddy, R.K., Gillilan, R.E., Cingolani, G.(2019) Biochemistry 58: 534-545
- PubMed: 30548067
- DOI: https://doi.org/10.1021/acs.biochem.8b01044
- Primary Citation of Related Structures:
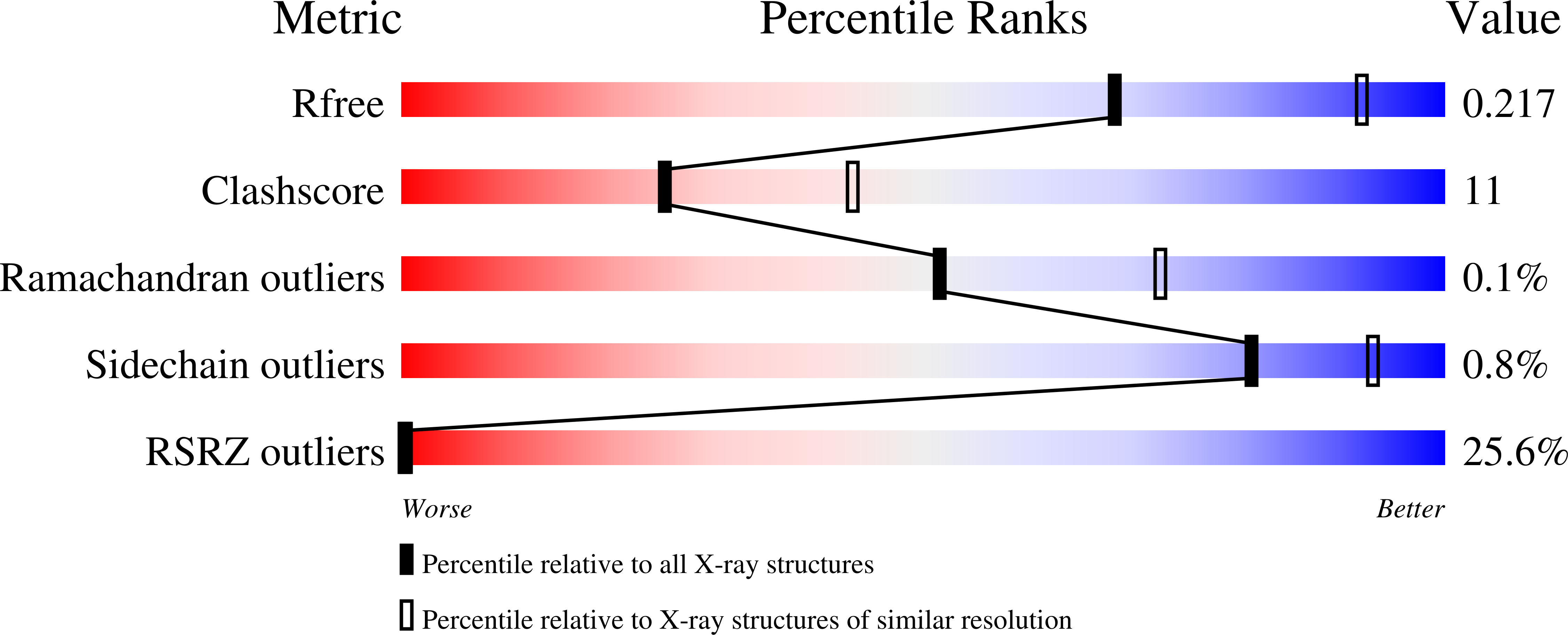
6E3B - PubMed Abstract:
Siw14 is a recently discovered inositol phosphatase implicated in suppressing prion propagation in Saccharomyces cerevisiae. In this paper, we used hybrid structural methods to decipher Siw14 molecular architecture. We found the protein exists in solution as an elongated monomer that is ∼140 Å in length, containing an acidic N-terminal domain and a basic C-terminal dual-specificity phosphatase (DSP) domain, structurally similar to the glycogen phosphatase laforin. The two domains are connected by a protease susceptible linker and do not interact in vitro. The crystal structure of Siw14-DSP reveals a highly basic phosphate-binding loop and an ∼10 Å deep substrate-binding crevice that evolved to dephosphorylate pyro-phosphate moieties. A pseudoatomic model of the full-length phosphatase generated from solution, crystallographic, biochemical, and modeling data sheds light on the interesting zwitterionic nature of Siw14, which we hypothesized may play a role in discriminating negatively charged inositol phosphates.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology , Thomas Jefferson University , 233 South 10th Street , Philadelphia , Pennsylvania 19107 , United States.