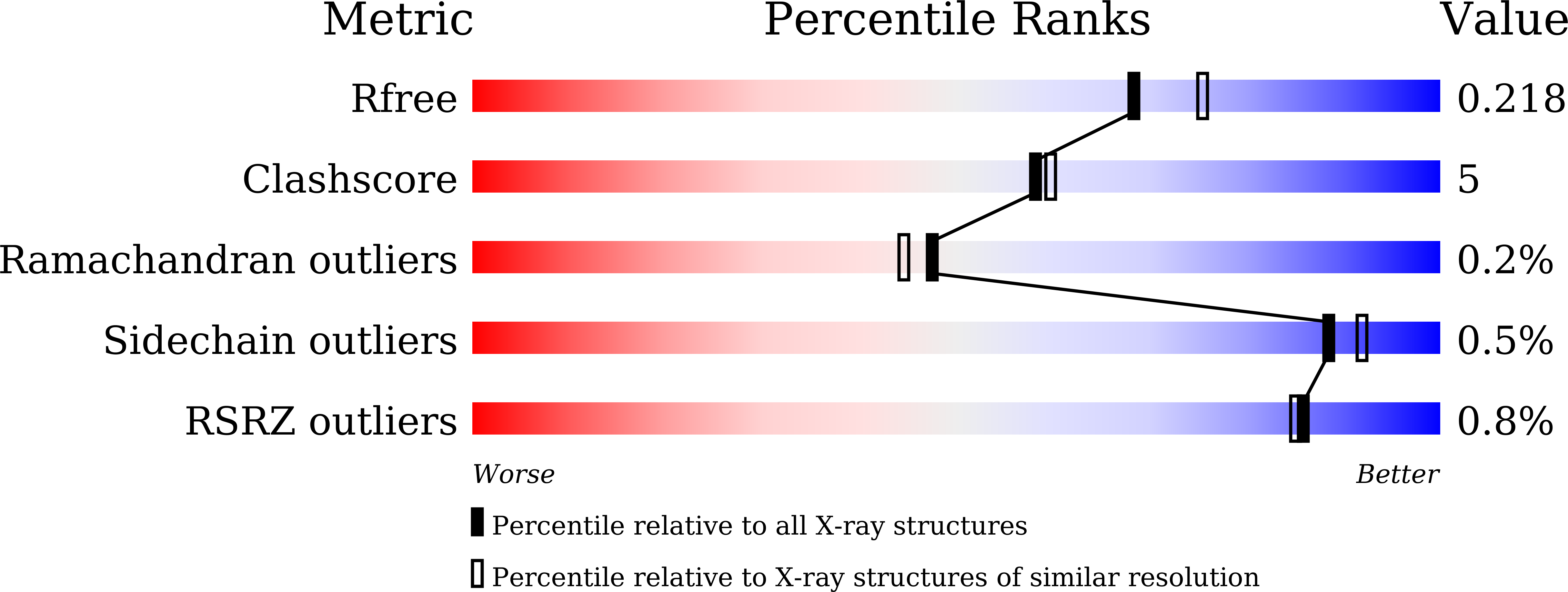
Structure of sirohydrochlorin ferrochelatase SirB: the last of the structures of the class II chelatase family.
Fujishiro, T., Shimada, Y., Nakamura, R., Ooi, M.(2019) Dalton Trans 48: 6083-6090
- PubMed: 30778451
- DOI: https://doi.org/10.1039/c8dt04727h
- Primary Citation of Related Structures:
5ZT7, 5ZT8 - PubMed Abstract:
The crystal structure of Bacillus subtilis SirB, which catalyses the insertion of Fe2+ into the substrate sirohydrochlorin (SHC) in siroheme biosynthesis, is reported herein as the last of the structures of class II chelatases. The structure of SirB with Co2+ showed that the active site of SirB is located at the N-terminal domain with metal-binding amino acid residues His10, Glu43, and His76, which was also predicted for CbiX, but is distinct from the C-terminal active sites of CbiK and HemH. The biosynthetic model reactions using SirB, Co2+ and uroporphyrin I or protoporphyrin IX as a SHC analogue revealed that SirB showed chelatase activity for uroporphyrin I, but not for protoporphyrin IX. Simulations of tetrapyrroles docking to SirB provided an insight into its tetrapyrrole substrate recognition: SHC and uroporphyrin I were suitably bound beside the Co2+ ion-binding site at the active site cavity; protoporphyrin IX was also docked to the active site but its orientation was different from those of the other two tetrapyrroles. Summarizing the present data, it was proposed that the key structural features for substrate recognition of SirB could be the hydrophobic area at the active site as well as the substituents of the tetrapyrroles.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Graduate School of Science and Engineering, Saitama University, 255 Shimo-Okubo, Sakura-ku, Saitama 338-8570, Japan. tfujishiro@mail.saitama-u.ac.jp.