Structure-based functional analysis of a PadR transcription factor from Streptococcus pneumoniae and characteristic features in the PadR subfamily-2.
Lee, C., Kim, M.I., Park, J., Oh, H., Kim, J., Hong, J., Jeon, B.Y., Ka, H., Hong, M.(2020) Biochem Biophys Res Commun 532: 251-257
- PubMed: 32868077
- DOI: https://doi.org/10.1016/j.bbrc.2020.08.035
- Primary Citation of Related Structures:
5ZQH - PubMed Abstract:
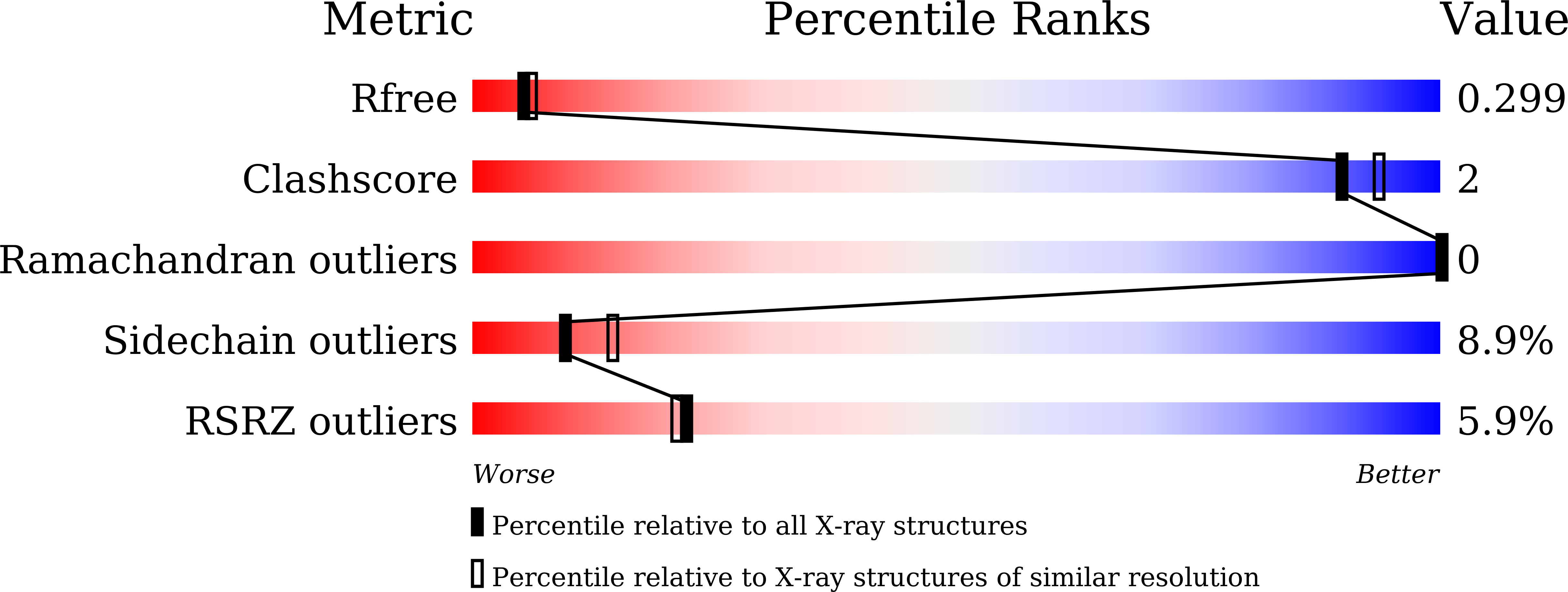
Since the first discovery of phenolic acid decarboxylase transcriptional regulator (PadR), its homologs have been identified mostly in bacterial species and constitute the PadR family. PadR family members commonly contain a winged helix-turn-helix (wHTH) motif and function as a transcription factor. However, the PadR family members are varied in terms of molecular size and structure. As a result, they are divided into PadR subfamily-1 and PadR subfamily-2. PadR subfamily-2 proteins have been reported in some pathogenic bacteria, including Listeria monocytogenes and Streptococcus pneumoniae, and implicated in drug resistance processes. Despite the growing numbers of known PadR family proteins and their critical functions in bacteria survival, biochemical and biophysical studies of the PadR subfamily-2 are limited. Here, we report the crystal structure of a PadR subfamily-2 member from Streptococcus pneumoniae (SpPadR) at a 2.40 Å resolution. SpPadR forms a dimer using its N-terminal and C-terminal helices. The two wHTH motifs of a SpPadR dimer expose their positively charged residues presumably to interact with DNA. Our structure-based mutational and biochemical study indicates that SpPadR specifically recognizes a palindromic nucleotide sequence upstream of its encoding region as a transcriptional regulator. Furthermore, comparative structural analysis of diverse PadR family members combined with a modeling study highlights the structural and regulatory features of SpPadR that are canonical to the PadR family or specific to the PadR subfamily-2.
Organizational Affiliation:
Division of Biological Science and Technology, Yonsei University, Wonju, 26493, Republic of Korea.