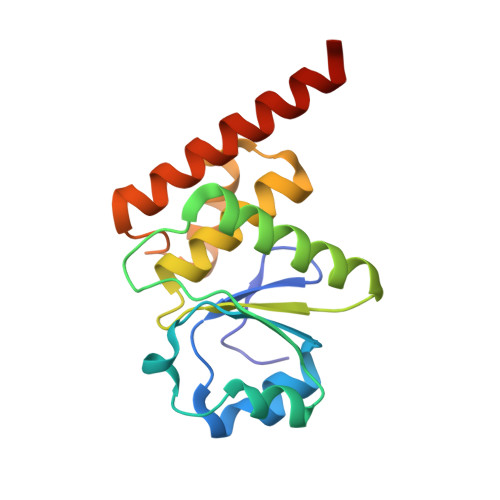
Structural study reveals the temperature-dependent conformational flexibility of Tk-PTP, a protein tyrosine phosphatase from Thermococcus kodakaraensis KOD1
Yun, H.Y., Lee, J., Kim, H., Ryu, H., Shin, H.C., Oh, B.H., Ku, B., Kim, S.J.(2018) PLoS One 13: e0197635-e0197635
- PubMed: 29791483
- DOI: https://doi.org/10.1371/journal.pone.0197635
- Primary Citation of Related Structures:
5Z59, 5Z5A, 5Z5B - PubMed Abstract:
Protein tyrosine phosphatases (PTPs) originating from eukaryotes or bacteria have been under intensive structural and biochemical investigation, whereas archaeal PTP proteins have not been investigated extensively; therefore, they are poorly understood. Here, we present the crystal structures of Tk-PTP derived from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1, in both the active and inactive forms. Tk-PTP adopts a common dual-specificity phosphatase (DUSP) fold, but it undergoes an atypical temperature-dependent conformational change in its P-loop and α4-α5 loop regions, switching between the inactive and active forms. Through comprehensive analyses of Tk-PTP, including additional structural determination of the G95A mutant form, enzymatic activity assays, and structural comparison with the other archaeal PTP, it was revealed that the presence of the GG motif in the P-loop is necessary but not sufficient for the structural flexibility of Tk-PTP. It was also proven that Tk-PTP contains dual general acid/base residues unlike most of the other DUSP proteins, and that both the residues are critical in its phosphatase activity. This work provides the basis for expanding our understanding of the previously uncharacterized PTP proteins from archaea, the third domain of living organisms.
Organizational Affiliation:
Disease Target Structure Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon, Republic of Korea.