Structure basis of the improved sweetness and thermostability of a unique double-sites single-chain sweet-tasting protein monellin (MNEI) mutant
Zhao, M., Xu, X., Liu, B.(2018) Biochimie 154: 156-163
- PubMed: 30195051
- DOI: https://doi.org/10.1016/j.biochi.2018.08.010
- Primary Citation of Related Structures:
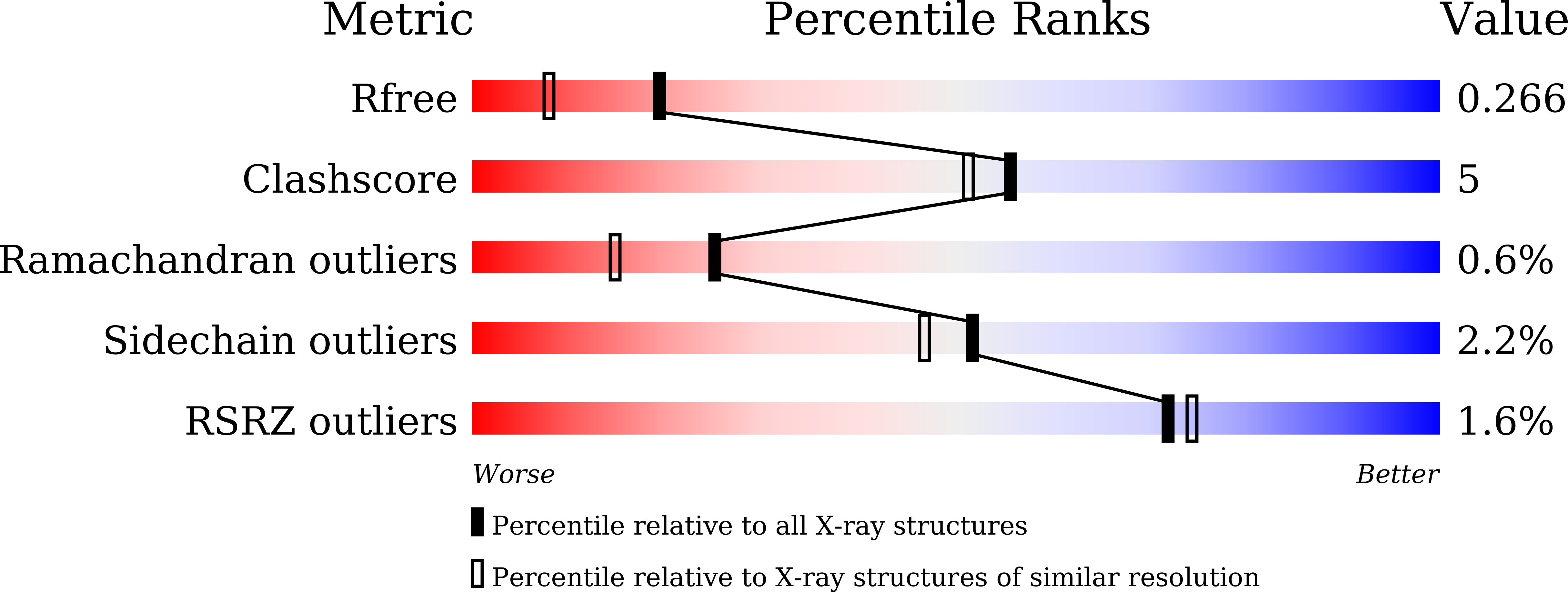
5Z1P - PubMed Abstract:
The sweet protein monellin has an intensely sweet potency but limited stability. We have identified a double-sites mutant (E2N/E23A) of the single-chain monellin (MNEI) with both improved sweetness (about 3-fold) and thermostability (10 °C). However, the structural basis of its superior properties remains elusive until now. Herein we report its crystal structure at a resolution 1.90 Å. Similar to the wild-type, E2N/E23A adopts a wedge-shaped structure consisting of a five-strand β-sheet partially "wrapped" around an α-helix. However, distinguishing parts were present in the loops region, including a remarkable conformation shift from β-strand to loop around residue R39. Molecular docking revealed the persistence of conserved protein-receptor interface and formation of new intermolecular ionic bonds in the E2N/E23A-receptor complex involving the taste-active residue R39 of the sweet protein, which could account for its significant improvement of sweetness. On the other hand, a rearrangement of intramolecular interaction network including the C-H … π bond between A23 and F89 that led to enhanced hydrophobicity in the protein core, could be correlated with its improved thermostability. Furthermore, two new sweeter mutants of MNEI were created. These findings highlight the critical roles of key sweetness determinant residue R39 and hydrophobicity at the protein core for the sweetness and thermostability of the protein, respectively, which thus provide a deeper insight for understanding the structure-function relationship of the sweet protein as well as guidance for rational design of this unique biomacromolecule.
Organizational Affiliation:
School of Food Science & Engineering, Qilu University of Technology (Shandong Academy of Sciences), No. 3501 University Road of Changqing District, 250353, Jinan, Shandong Province, China.