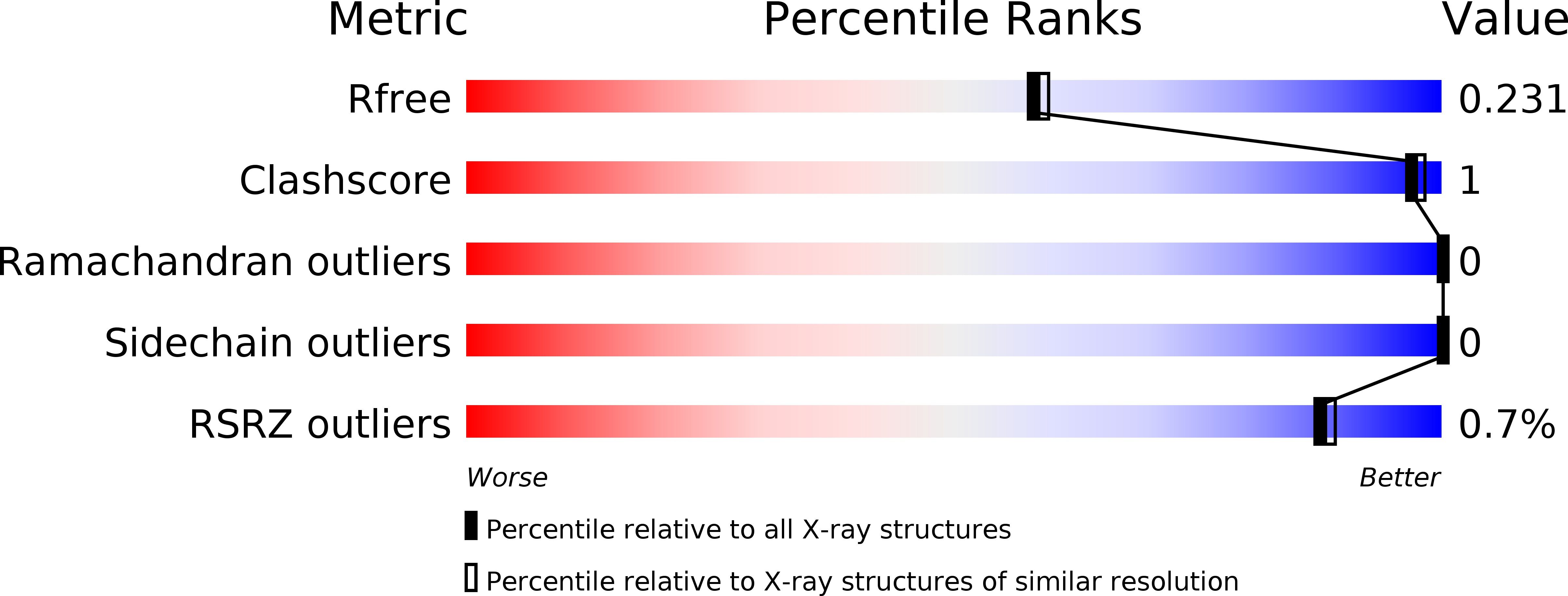
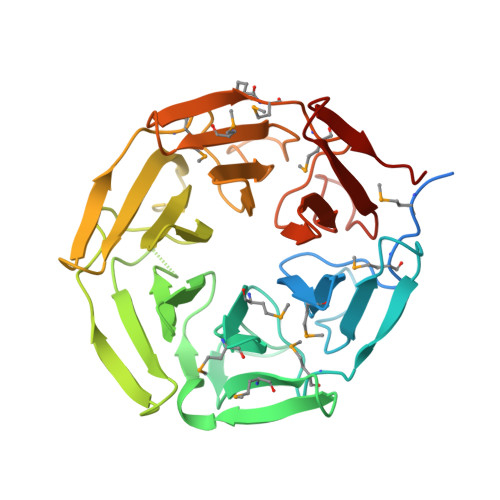
Crystal structure of the Kelch domain of human NS1-binding protein at 1.98 angstrom resolution.
Guo, L., Liu, Y.(2018) Acta Crystallogr F Struct Biol Commun 74: 174-178
- PubMed: 29497022
- DOI: https://doi.org/10.1107/S2053230X18001577
- Primary Citation of Related Structures:
5YY8 - PubMed Abstract:
NS1-binding protein (NS1-BP), which belongs to the Kelch protein superfamily, was first identified as a novel human 70 kDa protein that interacts with NS1 of Influenza A virus. It is involved in many cell functions, including pre-mRNA splicing, the ERK signalling pathway, the aryl hydrocarbon receptor (AHR) pathway, F-actin organization and protein ubiquitylation. However, the structure of NS1-BP is still unknown, which may impede functional studies. Here, the structure of the C-terminal Kelch domain of NS1-BP (NS1-BP-C; residues 330-642) was determined at 1.98 Å resolution. The Kelch domain adopts a highly symmetric six-bladed β-propeller fold structure. Each blade of the β-propeller is composed of four antiparallel β-strands. Comparison of the Kelch-domain structures of NS1-BP and its homologues showed that the Gly-Gly pair in β-strand B and the hydrophobic Trp residue in β-strand D are highly conserved, while the B-C loops in blades 2 and 6 are variable. This structure of the Kelch domain of NS1-BP extends the understanding of NS1-BP.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, People's Republic of China.