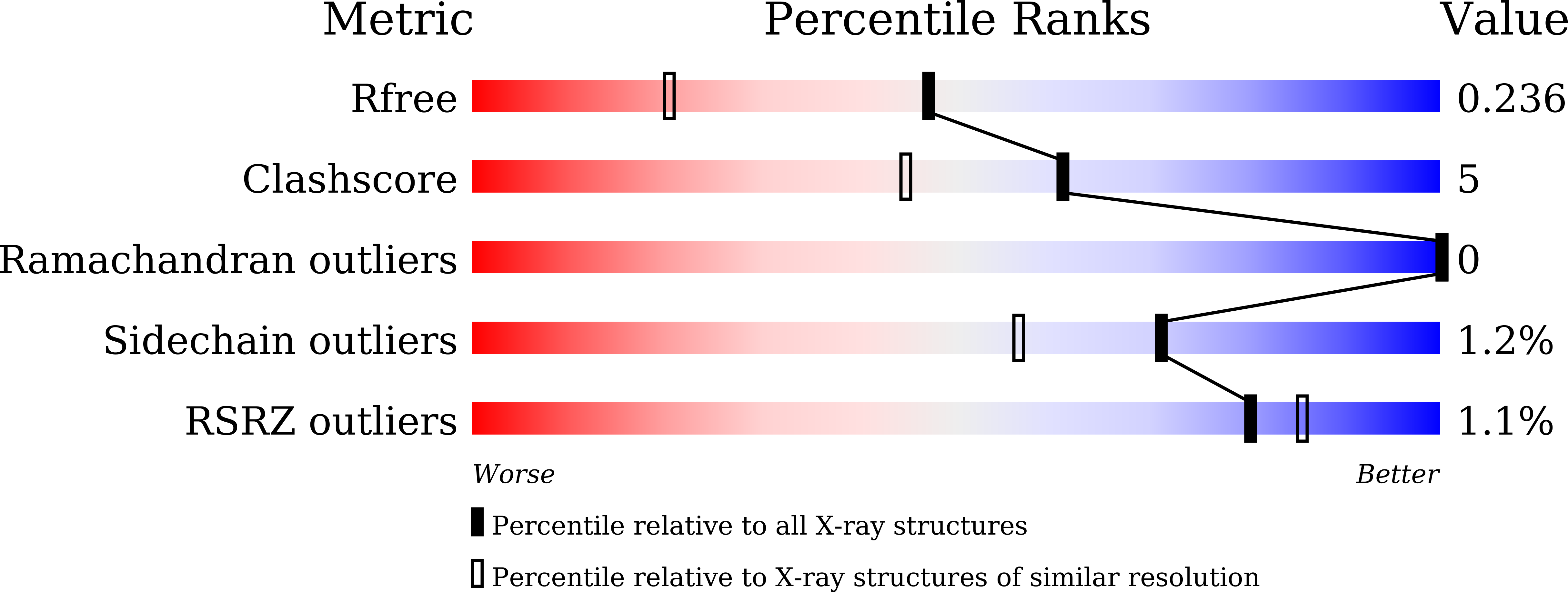
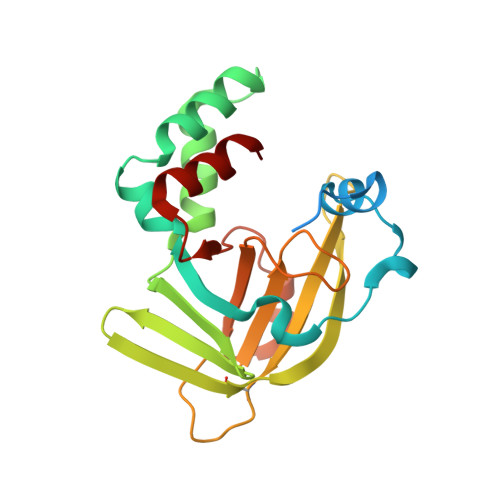
Crystal structure of E. coli ZinT with one zinc-binding mode and complexed with citrate
Chen, J., Wang, L., Shang, F., Dong, Y., Ha, N.C., Nam, K.H., Quan, C., Xu, Y.(2018) Biochem Biophys Res Commun 500: 139-144
- PubMed: 29596824
- DOI: https://doi.org/10.1016/j.bbrc.2018.03.192
- Primary Citation of Related Structures:
5YXC - PubMed Abstract:
The ZnuABC ATP-binding cassette transporter found in gram-negative bacteria has been implicated in ensuring adequate zinc import into Zn(II)-poor environments. ZinT is an essential component of ZnuABC and contributes to metal transport by transferring metals to ZnuA, which delivers them to ZnuB in periplasmic zinc recruitment. Although several structures of E. coli ZinT have been reported, its zinc-binding sites and oligomeric state have not been clearly identified. Here, we report the crystal structure of E. coli ZinT at 1.76 Å resolution. This structure contains one zinc ion in its calycin-like domain, and this ion is coordinated by three highly conserved histidine residues (His167, His176 and His178). Moreover, three oxygen atoms (O 1 , O 6 and O 7 ) from the citrate molecule interact with zinc, giving the zinc ion stable octahedral coordination. Our EcZinT structure shows the fewest zinc ions bound of all reported EcZinT structures. Crystallographic packing and size exclusion chromatography suggest that EcZinT prefers to form monomers in solution. Our results provide insights into the molecular function of ZinT.
Organizational Affiliation:
Department of Bioengineering, College of Life Science, Dalian Minzu University, Dalian 116600, Liaoning, China; Key Laboratory of Biotechnology and Bioresources Utilization (Dalian Minzu University), Ministry of Education, China.