The crystal structure of the Pyrococcus abyssi mono-functional methyltransferase PaTrm5b
Wu, J., Jia, Q., Wu, S., Zeng, H., Sun, Y., Wang, C., Ge, R., Xie, W.(2017) Biochem Biophys Res Commun 493: 240-245
- PubMed: 28911863
- DOI: https://doi.org/10.1016/j.bbrc.2017.09.038
- Primary Citation of Related Structures:
5YAC - PubMed Abstract:
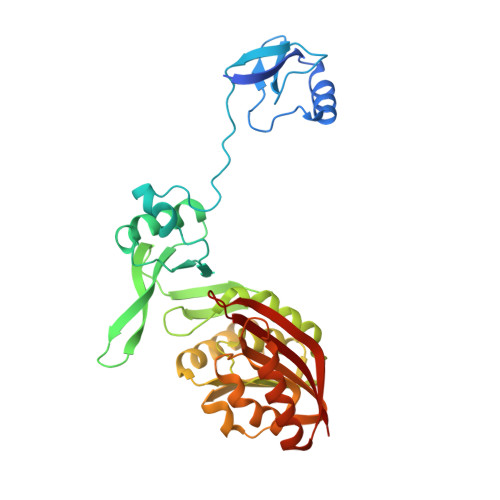
The wyosine hypermodification found exclusively at G37 of tRNA Phe in eukaryotes and archaea is a very complicated process involving multiple steps and enzymes, and the derivatives are essential for the maintenance of the reading frame during translation. In the archaea Pyrococcus abyssi, two key enzymes from the Trm5 family, named PaTrm5a and PaTrm5b respectively, start the process by forming N1-methylated guanosine (m 1 G37). In addition, PaTrm5a catalyzes the further methylation of C7 on 4-demethylwyosine (imG-14) to produce isowyosine (imG2) at the same position. The structural basis of the distinct methylation capacities and possible conformational changes during catalysis displayed by the Trm5 enzymes are poorly studied. Here we report the 3.3 Å crystal structure of the mono-functional PaTrm5b, which shares 32% sequence identity with PaTrm5a. Interestingly, structural superposition reveals that the PaTrm5b protein exhibits an extended conformation similar to that of tRNA-bound Trm5b from Methanococcus jannaschii (MjTrm5b), but quite different from the open conformation of apo-PaTrm5a or well folded apo-MjTrm5b reported previously. Truncation of the N-terminal D1 domain leads to reduced tRNA binding as well as the methyltransfer activity of PaTrm5b. The differential positioning of the D1 domains from three reported Trm5 structures were rationalized, which could be attributable to the dissimilar inter-domain interactions and crystal packing patterns. This study expands our understanding on the methylation mechanism of the Trm5 enzymes and wyosine hypermodification.
Organizational Affiliation:
School of Life Sciences, The Sun Yat-Sen University, Guangzhou, Guangdong 510275, People's Republic of China.