Substrate Recognition by a Dual-Function P450 Monooxygenase GfsF Involved in FD-891 Biosynthesis
Miyanaga, A., Takayanagi, R., Furuya, T., Kawamata, A., Itagaki, T., Iwabuchi, Y., Kanoh, N., Kudo, F., Eguchi, T.(2017) Chembiochem 18: 2179-2187
- PubMed: 28869713
- DOI: https://doi.org/10.1002/cbic.201700429
- Primary Citation of Related Structures:
5Y1I - PubMed Abstract:
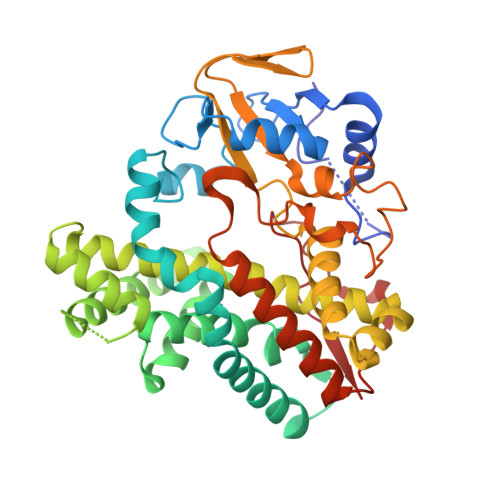
GfsF is a multifunctional P450 monooxygenase that catalyzes epoxidation and subsequent hydroxylation in the biosynthesis of macrolide polyketide FD-891. Here, we describe the biochemical and structural analysis of GfsF. To obtain the structural basis of a dual-function reaction, we determined the crystal structure of ligand-free GfsF, which revealed GfsF to have a predominantly hydrophobic substrate binding pocket. The docking models, in conjunction with the results of the enzymatic assay with substrate analogues and site-directed mutagenesis suggested two distinct substrate binding modes for epoxidation and hydroxylation reactions, which explained how GfsF regulates the order of two oxidative reactions. These findings provide new insights into the reaction mechanism of multifunctional P450 monooxygenases.
Organizational Affiliation:
Department of Chemistry, Tokyo Institute of Technology, 2-12-1 O-okayama, Meguro-ku, Tokyo, 152-8551, Japan.